Automating Exploratory Data Analysis via Machine Learning: An Overview


New Citation Alert added!
This alert has been successfully added and will be sent to:
You will be notified whenever a record that you have chosen has been cited.
To manage your alert preferences, click on the button below.
New Citation Alert!
Please log in to your account
Information & Contributors
Bibliometrics & citations.
- Mądziel M (2024) Quantifying Emissions in Vehicles Equipped with Energy-Saving Start–Stop Technology: THC and NOx Modeling Insights Energies 10.3390/en17122815 17 :12 (2815) Online publication date: 7-Jun-2024 https://doi.org/10.3390/en17122815
- Aldoseri A Al-Khalifa K Hamouda A (2024) Methodological Approach to Assessing the Current State of Organizations for AI-Based Digital Transformation Applied System Innovation 10.3390/asi7010014 7 :1 (14) Online publication date: 8-Feb-2024 https://doi.org/10.3390/asi7010014
- da Silva R Benso M Corrêa F Messias T Mendonça F Marques P Duarte S Mendiondo E Delbem A Saraiva A (2024) A Data-Driven Method for Water Quality Analysis and Prediction for Localized Irrigation AgriEngineering 10.3390/agriengineering6020103 6 :2 (1771-1793) Online publication date: 18-Jun-2024 https://doi.org/10.3390/agriengineering6020103
- Show More Cited By
Index Terms
Mathematics of computing
Probability and statistics
Statistical paradigms
Exploratory data analysis
Recommendations
Dataprep.eda: task-centric exploratory data analysis for statistical modeling in python.
Exploratory Data Analysis (EDA) is a crucial step in any data science project. However, existing Python libraries fall short in supporting data scientists to complete common EDA tasks for statistical modeling. Their API design is either too low level, ...
Automatically Generating Data Exploration Sessions Using Deep Reinforcement Learning
Exploratory Data Analysis (EDA) is an essential yet highly demanding task. To get a head start before exploring a new dataset, data scientists often prefer to view existing EDA notebooks -- illustrative, curated exploratory sessions, on the same dataset,...
A Space-Filling Multidimensional Visualization (SFMDVis for Exploratory Data Analysis
We introduce a new Space-Filling Multidimensional Data Visualization (SFMDVis) that can be used to facilitate the viewing, interaction and analysis of the multidimensional data with a fully utilized display space. The existing multidimensional ...
Information
Published in.

- General Chairs:
Portland State University, USA
University of British Columbia, Canada
- Program Chairs:
University of Wisconsin, USA
Megagon Labs, USA
- Publications Chairs:
University of Illinois at Urbana-Champaign, USA
RelationalAI, USA
- SIGMOD: ACM Special Interest Group on Management of Data
Association for Computing Machinery
New York, NY, United States
Publication History
Permissions, check for updates, author tags.
- data exploration
- exploratory data analysis
- Short-paper
Funding Sources
- Israel Innovation Authority
- Israel Science Foundation
- Len Blavatnik and the Blavatnik Family foundation
- United States-Israel Binational Science Foundation
Acceptance Rates
Contributors, other metrics, bibliometrics, article metrics.
- 37 Total Citations View Citations
- 1,638 Total Downloads
- Downloads (Last 12 months) 441
- Downloads (Last 6 weeks) 39
- Sinap V (2024) Perakende Sektöründe Makine Öğrenmesi Algoritmalarının Karşılaştırmalı Performans Analizi: Black Friday Satış Tahminlemesi Selçuk Üniversitesi Sosyal Bilimler Meslek Yüksekokulu Dergisi 10.29249/selcuksbmyd.1401822 27 :1 (65-90) Online publication date: 30-Apr-2024 https://doi.org/10.29249/selcuksbmyd.1401822
- Tsai Y (2024) Empowering students through active learning in educational big data analytics Smart Learning Environments 10.1186/s40561-024-00300-1 11 :1 Online publication date: 1-Apr-2024 https://doi.org/10.1186/s40561-024-00300-1
- Zhu J Cai P Niu B Ni Z Xu K Huang J Wan J Ma S Wang B Zhang D Tang L Liu Q (2024) Chat2Query: A Zero-Shot Automatic Exploratory Data Analysis System with Large Language Models 2024 IEEE 40th International Conference on Data Engineering (ICDE) 10.1109/ICDE60146.2024.00420 (5429-5432) Online publication date: 13-May-2024 https://doi.org/10.1109/ICDE60146.2024.00420
- Xiang S Deng H Wu J Liu J (2024) Exploring the Integration of Artificial Intelligence in Research Processes of Graduate Students 2024 6th International Conference on Computer Science and Technologies in Education (CSTE) 10.1109/CSTE62025.2024.00027 (110-113) Online publication date: 19-Apr-2024 https://doi.org/10.1109/CSTE62025.2024.00027
- Yue J Tu M Leong Y (2024) A spatial analysis of the health and longevity of Taiwanese people The Geneva Papers on Risk and Insurance - Issues and Practice 10.1057/s41288-024-00322-3 49 :2 (384-399) Online publication date: 4-Apr-2024 https://doi.org/10.1057/s41288-024-00322-3
- Moral P Mustafi D Sahana S (2024) PODBoost: an explainable AI model for polycystic ovarian syndrome detection using grey wolf-based feature selection approach Neural Computing and Applications 10.1007/s00521-024-10171-9 Online publication date: 30-Jul-2024 https://doi.org/10.1007/s00521-024-10171-9
- Vázquez-Ingelmo A García-Holgado A Pozo-Zapatero J (2024) A Proposal for an Interactive Assistant to Support Exploratory Data Analysis in Educational Settings Learning and Collaboration Technologies 10.1007/978-3-031-61685-3_20 (272-281) Online publication date: 1-Jun-2024 https://doi.org/10.1007/978-3-031-61685-3_20
View Options
Login options.
Check if you have access through your login credentials or your institution to get full access on this article.
Full Access
View options.
View or Download as a PDF file.
View online with eReader .
Share this Publication link
Copying failed.
Share on social media
Affiliations, export citations.
- Please download or close your previous search result export first before starting a new bulk export. Preview is not available. By clicking download, a status dialog will open to start the export process. The process may take a few minutes but once it finishes a file will be downloadable from your browser. You may continue to browse the DL while the export process is in progress. Download
- Download citation
- Copy citation
We are preparing your search results for download ...
We will inform you here when the file is ready.
Your file of search results citations is now ready.
Your search export query has expired. Please try again.
This week: the arXiv Accessibility Forum
Help | Advanced Search
Computer Science > Human-Computer Interaction
Title: goals, process, and challenges of exploratory data analysis: an interview study.
Abstract: How do analysis goals and context affect exploratory data analysis (EDA)? To investigate this question, we conducted semi-structured interviews with 18 data analysts. We characterize common exploration goals: profiling (assessing data quality) and discovery (gaining new insights). Though the EDA literature primarily emphasizes discovery, we observe that discovery only reliably occurs in the context of open-ended analyses, whereas all participants engage in profiling across all of their analyses. We describe the process and challenges of EDA highlighted by our interviews. We find that analysts must perform repetitive tasks (e.g., examine numerous variables), yet they may have limited time or lack domain knowledge to explore data. Analysts also often have to consult other stakeholders and oscillate between exploration and other tasks, such as acquiring and wrangling additional data. Based on these observations, we identify design opportunities for exploratory analysis tools, such as augmenting exploration with automation and guidance.
| Subjects: | Human-Computer Interaction (cs.HC) |
| Cite as: | [cs.HC] |
| (or [cs.HC] for this version) | |
| Focus to learn more arXiv-issued DOI via DataCite |
Submission history
Access paper:.
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
DBLP - CS Bibliography
Bibtex formatted citation.
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
Identifying the Steps in an Exploratory Data Analysis: A Process-Oriented Approach
- Conference paper
- Open Access
- First Online: 26 March 2023
- Cite this conference paper
You have full access to this open access conference paper

- Seppe Van Daele 9 &
- Gert Janssenswillen ORCID: orcid.org/0000-0002-7474-2088 9
Part of the book series: Lecture Notes in Business Information Processing ((LNBIP,volume 468))
Included in the following conference series:
- International Conference on Process Mining
4596 Accesses
Best practices in (teaching) data literacy, specifically Exploratory Data Analysis, remain an area of tacit knowledge until this day. However, with the increase in the amount of data and its importance in organisations, analysing data is becoming a much-needed skill in today’s society. Within this paper, we describe an empirical experiment that was used to examine the steps taken during an exploratory data analysis, and the order in which these actions were taken. Twenty actions were identified. Participants followed a rather iterative process of working step by step towards the solution. In terms of the practices of novice and advanced data analysts, few relevant differences were yet discovered.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others

Ways of Working in the Interpretive Tradition
A practical, iterative framework for secondary data analysis in educational research

Questions as data: illuminating the potential of learning analytics through questioning an emergent field
- Process mining
- Deliberate practice
- Learning analytics
1 Introduction
Data is sometimes called the new gold, but is much better compared to gold-rich soil. As with gold mining, several steps are needed to go through in order to get to the true value. With the amount and importance of data in nearly every industry [ 13 , 14 , 15 ], data analysis is a vital skill in the current job market, not limited to profiles such as data scientists or machine learning engineers, but equally important for marketing analysts, business controllers, as well as sport coaches, among others.
However, best practices in data literacy, and how to develop them, mainly remains an area of tacit knowledge until this day, specifically in the area of Exploratory Data Analysis (EDA). EDA is an important part in the data analysis process where interaction between the analyst and the data is high [ 3 ]. While there are guidelines on how the process of data analysis can best be carried out [ 15 , 18 , 21 ], these steps typically describe what needs to be done at a relatively high level, and do not precisely tell how best to perform them in an actionable manner. Which specific steps take place during an exploratory data analysis, and how they are structured in an analysis has not been investigated.
The goal of this paper is to refine the steps underlying exploratory data analysis beyond high-level categorisations such as transforming, visualising, and modelling. In addition, we analyse the order in which these actions are performed. The results of this paper form a first step towards better understanding the detailed steps in a data analysis, which can be used in future research to analyse difference between novices and experts in data analysis, and create better data analysis teaching methods focussed on removing these differences.
The next section will discuss related work, while Sect. 3 will discuss the methodology used. The identified steps are described in the subsequent section, while an analysis of the recorded data is provided in Sect. 5 . Section 6 concludes the paper.
2 Related Work
A number of high-level tasks to be followed while performing a data analysis have already been defined in the literature [ 15 , 18 ], which can be synthesised as 1) the collection of data, 2) processing of data, 3) cleaning of data, 4) exploratory data analysis, 5) predictive data analysis, and 6) communicating the results. In [ 21 ] this process is elaborated in more detail, applied to the R language. Here the process starts with importing data and cleaning. The actual data analysis is subsequently composed of the cycle of transforming, visualising and modelling data, and is thus slightly more concrete than the theoretical exploratory and prescriptive data analysis. The concluding communication step is similar to [ 15 , 18 ].
That the different steps performed in a data analysis have received little attention, has also been recognised by [ 23 ], specifically focused on process analysis. In this paper, an empirical study has been done to understand how process analysts follow different patterns in analysing process data, and have different strategies to explore event data. Subsequent research has shown that such analysis can lead to the identification of challenges to improve best practices [ 24 ].
Breaking down a given action into smaller steps can reduce cognitive load when performing the action [ 20 ]. Cognitive load is the load that occurs when processing information. The more complex this information is, the higher the cognitive load is. Excessive cognitive load can overload working memory and thus slow down the learning process. Creating an instruction manual addresses The Isolated Elements Effect [ 4 ], when there is a reduction in cognitive load by isolating steps, and only then looking at the bigger picture [ 20 ]. In [ 5 ], this theory was applied using The Structured Process Modeling Theory , to reduce the cognitive load when creating a process model. Participants who followed structured steps, thus reducing their cognitive load, generally made fewer syntax errors and created better process models [ 5 ]. Similarly, in [ 10 ], participants were asked to build an event log, where the test group was provided with the event log building guide from [ 11 ]. The results showed that the event logs built by the test group outperformed those of the control group in several areas.
An additional benefit of identifying smaller steps is that these steps can be used in the creation of a deliberate practice —a training course that meets the following conditions [ 1 , 6 ] :
Tasks with a defined objective
Immediate feedback on the task created
Opportunity to repeat this task multiple times
Motivation to actually get better
Karl Ericsson [ 6 ] studied what the training of experts in different fields had in common [ 2 ], from which the concept of deliberate practice emerged. It was already successfully applied, for example, in [ 7 ] where a physics course, reworked to deliberate practise principles, resulted in higher attendance and better grades.
In addition to studying what kind of training experts use to acquire their expertise, it has also been studied why experts are better at a particular field than others. In [ 6 ], it is concluded that experts have more sophisticated mental representations that enable them to make better and/or faster decisions. Mental representations are internal models about certain information that become more refined with training [ 6 ]. Identifying actions taken in a data analysis can help in mapping mental representations of data analysis experts. This can be done by comparing the behaviour of experts with that of beginners. Knowing why an expert performs a certain action at a certain point can have a positive effect on the development of beginners’ mental models. In fact, using mental representations of experts was considered in [ 19 ] as a crucial first step in designing new teaching methods.
3 Methodology
In order to analyse the different steps performed during an exploratory analysis, and typical flows between them, an experiment was conducted. The experiments and further data processing and analysis steps are described below.
Experiment. Cognitive Task Analysis (CTA) [ 22 ] was used as overall methodology for conducting the experiment described in this paper, with the aim to uncover (hidden) steps in a participant’s process of exploratory data analysis. Participants were asked to make some simple analyses using supplied data and to make a screen recording of this process. The tasks concerned analysing the distribution of variables, the relationship between variables, as well as calculating certain statistics.
As certain steps can be taken for granted due to developed automatisms [ 8 ], the actual analysis was followed by an interview, in which the participants were asked to explain step by step what decisions and actions were taken. By having the interview take place after the data analysis, interference with the participants’ usual way of working was avoided. For example, asking questions before or during the data analysis could have caused participants to hesitate, slow down, or even make different choices.
The general structure of the experiment was as follows:
Participants: The participants for this experiment were invited by mail from three groups with different levels of experience: undergraduate students, graduate students, and PhD students, from the degree Business and Information systems engineering. These students received an introductory course on data analysis in their first bachelor year, where they work with the language \(\textrm{R}\) , which was subsequently chosen as the language to be used in the experiment. In the end, 11 students were convinced to participate in this experiment: two undergraduate students, four graduate students and 5 PhD students. The 11 participants each performed the complete analysis of three assignments, and thus results from 33 assignments were collected. While having participants with different levels of experience is expected to result in a broader variety in terms of behaviour, the scale of the experiment and the use of student participants only will not allow a detailed analysis of the relationship between experience-level and analysis behaviour. Furthermore, disregarding the different level of students, the once accepting the invitation to participate might also be the more confident about their skills.
Survey: Before participants began the data analysis, they were asked to complete an introductory survey to gain insight into their own perceptions of their data analysis skill (in \(\textrm{R}\) ).
Assignment: The exploratory analysis was done in the R programming language, and consisted of three independent tasks about data from a housing market: 2 involving data visualisation and 1 specific quantitative question. The analysis was recorded by the participants.
Interview: The recording of the assignment was used during the interview to find out what steps, according to the participants themselves, were taken. Participants were asked to actively tell what actions were taken and why.
Transcription. The transcription of the interviews was done manually. Because most participants actively narrated the actions taken, a question-answer structure was not chosen. If a question was still asked, it was placed in italics between two dashes when transcribed.
Coding and Categorization. To code the transcripts of the interviews, a combination of descriptive and process coding was used in the first iteration. Descriptive coding looks for nouns that capture the content of the sentence [ 16 ]. Process coding, in turn, attempts to capture actions by encoding primarily action-oriented words (verbs) [ 16 ]. These coding techniques were applied to the transcripts by highlighting the words and sentences that met them. A second iteration used open coding (also known as initial coding) where the marked codes from the first iteration were grouped with similarly marked codes [ 9 , 17 ]. These iterations were performed one after the other for the same transcription before starting the next transcription.
These resulting codes were the input for constructing the categories. In this process, the codes that had the same purpose were taken together and codes with a similar purpose were grouped together and given an overarching term. This coding step is called axial coding [ 9 ].
Event Log Construction. Based on the screen recording and the transcription, the actions found were transformed into an event log. In addition, if applicable, additional information was also stored to enrich the data such as the location where a certain action was performed (e.g. in the console, in a script, etc.), what exactly happened in the action (e.g. what was filtered on) and then an attribute how this happened (e.g. search for a variable using CTRL+F ). Timestamps for the event log where based on the screen recordings.
Event Log Analysis. The frequency of activities, and typical activity flows were subsequently analysed. Next to the recorded behaviour, also the quality of the execution was assessed, by looking at both the duration of the analysis, as well as the correctness of the results. For each of these focus points, participants with differing levels of experiences where also compared.
For the analysis of the event log, the \(\textrm{R}\) package bupaR was used [ 12 ]. Because there were relatively few cases present in the event log, the analysis also consisted largely of qualitative analysis of the individual traces.
4 Identified Actions
Before analysing the executed actions and flows in relation to the different experiences, duration and correctness, this section describes the identified actions, which have been subdivided in the categories preparatory, analysis, debugging, and other actions.
Preparatory Actions. Actions are considered preparatory steps if they occurred mainly prior to the actual analysis itself. For the purpose of this experiment, actions were selected that had a higher relative frequency among the actions performed before the first question than during the analysis. An overview of preparatory actions is shown in Table 1 .
Analysis Actions. The steps covered within this category are actions that can be performed to accomplish a specific task, and are listed in Table 2 . These are actions directly related to solving the data analysis task and not, for example, emergency actions that must be performed such as solving an error message.
Debugging Actions. Debugging is the third category of operations that was identified. Next to the actual debugging of the code, this category include the activities that (might) trigger debugging, which are errors , warnings , and messages .
Executing the code 77 times out of 377 resulted in an error. Debugging is a (series of) action(s) taken after receiving an error or warning. Most of these errors were fairly trivial to resolve. In twenty percent of the loglines registered during debugging, however, additional information was consulted on, for example, the Internet.
Other Actions. The last category of actions includes adding structure, reasoning, reviewing the assignments, consulting information, and trial-and-error. Except for the review of the assignments, which was performed after completing all the assignments, these actions are fairly independent of the previous action and thus were performed at any point in the analysis. An overview of these actions can be found in Table 3 . Note that as trial-and-error is a method rather than a separate action, it was not coded separately in the event log, but can be identified in the log as a pattern.
In the experiment, a total of 1674 activity instances were recorded. An overview of the identified actions together with summary statistics is provided in Table 4 . It can be seen that the most often observed actions are Execute code , Consult information , Prepare data and Evaluate results . Twelve of the identified actions were performed by all 11 participants at some point. Looking at the summary statistics, we observe quite significant differences in the execution frequency of actions, such as the consultation of information (ranging from 4 to 63) and the execution of code (ranging from 16 to 48), indicating important individual differences. Table 5 shows for each participant the total processing time (minutes) together with the total number of actions, and the number of actions per category.
Flows. A first observation is that the log records direct repetitions of a certain number of actions. This is a natural consequence of the fact that information is stored in additional attributes. As such, when a participant is, for instance, consulting different sources of information directly after one another, this will not be regarded as a single “Consulting information” action, but as a sequence of smaller actions. Information of these repetitions is shown in Table 6 . Because these length-one loops might clutter the analysis, it was decided to collapse them into single activity instances. After doing so, the number of activity instances was reduced from 1674 to 1572.
That the process of data analysis is flexible attests Fig. 1 , which contains a directly-follows matrix of the log. While many different (and infrequent) flows can be observed, some interesting insights can be seen. Within the analysis actions, we can see 2 groups: actions related to manipulation of data, and actions related to evaluation and visualising data. Furthermore, it can be seen that some analysis actions are often performed before or after preparatory actions, while most are not.
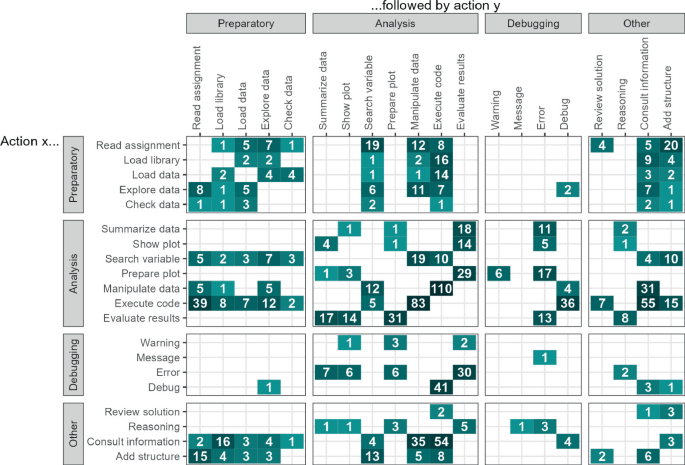
Precedence flows between actions.
Duration. In Fig. 2 , the total time spent on each of the 4 categories is shown per participant, divided in undergraduate, graduate and PhD participants. The dotted vertical lines in each group indicates the average time spent. While the limited size of the experiment does not warrant generalizable results with respect to different experience levels, it can be seen that Undergraduates spent the least time overall, while graduate spent the most time. In the latter group, we can however see a large amount of variation between participants. What is notable is that both graduate participants and PhDs spent a significantly larger amount of time on preparatory steps, compared to undergraduate students. On average, graduate students spent more time on other actions than the other groups. Predominantly, this appeared to be the consultation of information. This might be explained by the fact that for these students, data analysis (specifically the course in R) was further removed in the past compared to undergraduate students. On the other hand, PhDs might have more expertise about usage of R and data analysis readily available.
Correctness. After the experiment, the results where also scored for correctness. Table 7 shows the average scores in each group, on a scale from zero to 100%. While the differences are small, and still noting the limited scope of the experiment, a slight gap can be observed between undergraduates on the one hand, and PhDs and graduates on the other. The gap between the latter two is less apparent.
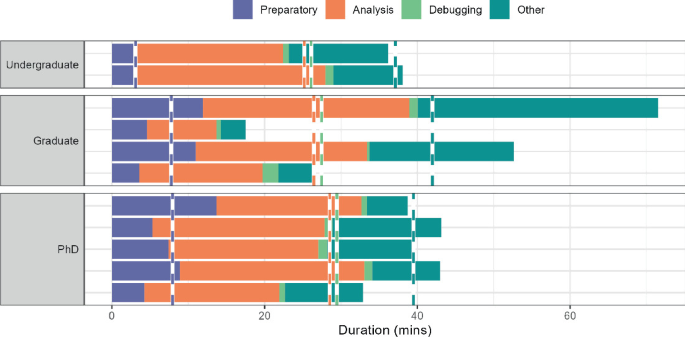
Duration per category for each participant in each experience level.
Figure 3 shows a correlation matrix between the scores, the number of actions in each category, and the time spent on each category. Taking into account the small data underlying these correlations, it can be seen that no significant positive correlations with the score can be observed. However, the score is found to have a moderate negative correlation with both the amount and duration of debugging actions, as well as the duration of analysis actions. While the former seems logical, the latter is somewhat counter-intuitive. Given that no relation is found between with the number of analysis actions, the average duration of an analysis task seems to relevant. This might thus indicate that the score is negatively influenced when the analysis takes place slower, which might be a sign of inferior skills.
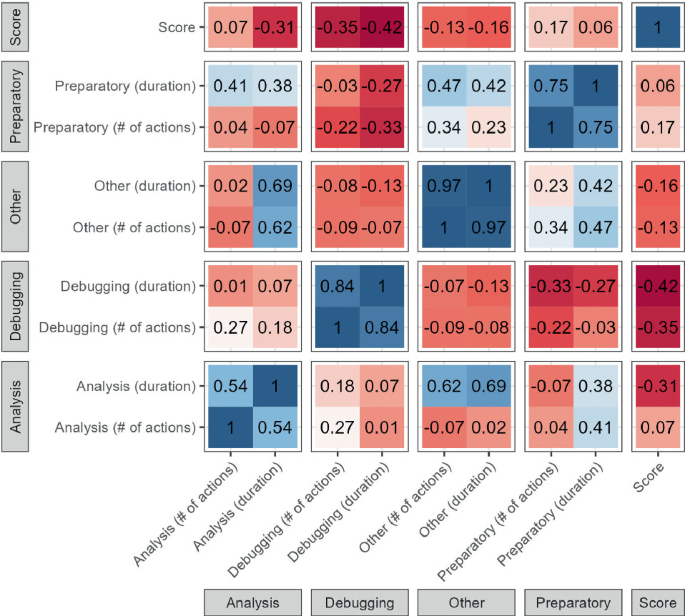
Correlations between score, number of (distinct) actions in each category, and duration of each category.
6 Conclusion
The steps completed during an exploratory data analysis can be divided into four categories: the preparatory steps, the analysis steps, the debug step, and finally the actions that do not belong to a category but can be used throughout the analysis process. By further breaking down the exploratory data analysis into these steps, it becomes easier to proceed step by step and thus possibly obtain better analyses. The data analysis process performed by the participants appeared to be an iterative process that involved working step-by-step towards the solution.
The experiment described in this paper clearly is only a first step towards understanding the behaviour of data analysts. Only a small amount of people participated and the analysis requested was a relatively simple exercise. As a result, the list of operations found might not be exhaustive. Furthermore, the use of R and RStudio will have caused that some of the operations are specifically related to R. While R was chosen because all participants had a basic knowledge of R through an introductory course received in the first bachelor year, future research is needed to see whether these steps are also relevant with respect to other programming languages or tools. Moreover, this course may have already taught a certain methodology, which might not generalize to other data analyst. Additionally, the fact that the participants participated voluntarily, might mean they feel more comfortable performing a data analysis in \(\textrm{R}\) than their peers, especially among novices.
It is recommended that further research is conducted on both the operations, the order of these operations as well as the practices of experts and novices. By using more heterogeneous participants, a more difficult task and different programming languages, it is expected that additional operations can be identified as well as differences in practices between experts and beginners. These can be used to identify the mental representations of experts and, in turn, can be used to design new teaching methods [ 19 ]. In addition, an analysis at the sub-activity level could provide insights about frequencies and a lower-level order, such as in what order the sub-activities in the act of preparing data were usually performed.
Anders Ericsson, K.: Deliberate practice and acquisition of expert performance: a general overview. Acad. Emerg. Med. 15 (11), 988–994 (2008)
Article Google Scholar
Anders Ericsson, K., Towne, T.J.: Expertise. Wiley Interdisc. Rev.: Cogn. Sci. 1 (3), 404–416 (2010)
Behrens, J.T.: Principles and procedures of exploratory data analysis. Psychol. Methods 2 (2), 131 (1997)
Blayney, P., Kalyuga, S., Sweller, J.: Interactions between the isolated-interactive elements effect and levels of learner expertise: experimental evidence from an accountancy class. Instr. Sci. 38 (3), 277–287 (2010)
Claes, J., Vanderfeesten, I., Gailly, F., Grefen, P., Poels, G.: The structured process modeling theory (SPMT) a cognitive view on why and how modelers benefit from structuring the process of process modeling. Inf. Syst. Front. 17 (6), 1401–1425 (2015). https://doi.org/10.1007/s10796-015-9585-y
Ericsson, A., Pool, R.: Peak: Secrets from the New Science of Expertise. Random House, New York (2016)
Google Scholar
Ericsson, K.A., et al.: The influence of experience and deliberate practice on the development of superior expert performance. Cambridge Handb. Expertise Expert Perform. 38 (685–705), 2–2 (2006)
Hinds, P.J.: The curse of expertise: the effects of expertise and debiasing methods on prediction of novice performance. J. Exp. Psychol. Appl. 5 (2), 205 (1999)
Holton, J.A.: The coding process and its challenges. Sage Handb. Grounded Theory 3 , 265–289 (2007)
Jans, M., Soffer, P., Jouck, T.: Building a valuable event log for process mining: an experimental exploration of a guided process. Enterp. Inf. Syst. 13 (5), 601–630 (2019)
Jans, M.: From relational database to valuable event logs for process mining purposes: a procedure. Technical report, Hasselt University, Technical report (2017)
Janssenswillen, G., Depaire, B., Swennen, M., Jans, M., Vanhoof, K.: bupaR: enabling reproducible business process analysis. Knowl.-Based Syst. 163 , 927–930 (2019)
Kitchin, R.: The Data Revolution: Big Data, Open Data, Data Infrastructures and their Consequences. Sage, New York (2014)
Mayer-Schoenberger, V., Cukier, K.: The rise of big data: how it’s changing the way we think about the world. Foreign Aff. 92 (3), 28–40 (2013)
O’Neil, C., Schutt, R.: Doing Data Science: Straight Talk from the Frontline. O’Reilly Media Inc, California (2013)
Saldaña, J.: Coding and analysis strategies. The Oxford handbook of qualitative research, pp. 581–605 (2014)
Saldaña, J.: The coding manual for qualitative researchers. The coding manual for qualitative researchers, pp. 1–440 (2021)
Saltz, J.S., Shamshurin, I.: Exploring the process of doing data science via an ethnographic study of a media advertising company. In: 2015 IEEE International Conference on Big Data (Big Data), pp. 2098–2105. IEEE (2015)
Spector, J.M., Ohrazda, C.: Automating instructional design: approaches and limitations. In: Handbook of Research on Educational Communications and Technology, pp. 681–695. Routledge (2013)
Sweller, J.: Cognitive load theory: Recent theoretical advances. (2010)
Wickham, H., Grolemund, G.: R for Data Science: Import, Tidy, Transform, Visualize, and Model Data. O’Reilly Media Inc, California (2016)
Yates, K.A., Clark, R.E.: Cognitive task analysis. International Handbook of Student Achievement. New York, Routledge (2012)
Zerbato, F., Soffer, P., Weber, B.: Initial insights into exploratory process mining practices. In: Polyvyanyy, A., Wynn, M.T., Van Looy, A., Reichert, M. (eds.) BPM 2021. LNBIP, vol. 427, pp. 145–161. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-85440-9_9
Chapter Google Scholar
Zimmermann, L., Zerbato, F., Weber, B.: Process mining challenges perceived by analysts: an interview study. In: Augusto, A., Gill, A., Bork, D., Nurcan, S., Reinhartz-Berger, I., Schmidt, R. (eds.) International Conference on Business Process Modeling, Development and Support, International Conference on Evaluation and Modeling Methods for Systems Analysis and Development, vol. 450, pp. 3–17. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-07475-2_1
Download references
Author information
Authors and affiliations.
Faculty of Business Economics, UHasselt - Hasselt University, Agoralaan, 3590, Diepenbeek, Belgium
Seppe Van Daele & Gert Janssenswillen
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Gert Janssenswillen .
Editor information
Editors and affiliations.
Free University of Bozen-Bolzano, Bozen-Bolzano, Italy
Marco Montali
York University, Toronto, ON, Canada
Arik Senderovich
Humboldt-Universität zu Berlin, Berlin, Germany
Matthias Weidlich
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Reprints and permissions
Copyright information
© 2023 The Author(s)
About this paper
Cite this paper.
Daele, S.V., Janssenswillen, G. (2023). Identifying the Steps in an Exploratory Data Analysis: A Process-Oriented Approach. In: Montali, M., Senderovich, A., Weidlich, M. (eds) Process Mining Workshops. ICPM 2022. Lecture Notes in Business Information Processing, vol 468. Springer, Cham. https://doi.org/10.1007/978-3-031-27815-0_38
Download citation
DOI : https://doi.org/10.1007/978-3-031-27815-0_38
Published : 26 March 2023
Publisher Name : Springer, Cham
Print ISBN : 978-3-031-27814-3
Online ISBN : 978-3-031-27815-0
eBook Packages : Computer Science Computer Science (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Warning: The NCBI web site requires JavaScript to function. more...
An official website of the United States government
The .gov means it's official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you're on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- Browse Titles
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-.

StatPearls [Internet].
Exploratory data analysis: frequencies, descriptive statistics, histograms, and boxplots.
Jacob Shreffler ; Martin R. Huecker .
Affiliations
Last Update: November 3, 2023 .
- Definition/Introduction
Researchers must utilize exploratory data techniques to present findings to a target audience and create appropriate graphs and figures. Researchers can determine if outliers exist, data are missing, and statistical assumptions will be upheld by understanding data. Additionally, it is essential to comprehend these data when describing them in conclusions of a paper, in a meeting with colleagues invested in the findings, or while reading others’ work.
- Issues of Concern
This comprehension begins with exploring these data through the outputs discussed in this article. Individuals who do not conduct research must still comprehend new studies, and knowledge of fundamentals in analyzing data and interpretation of histograms and boxplots facilitates the ability to appraise recent publications accurately. Without this familiarity, decisions could be implemented based on inaccurate delivery or interpretation of medical studies.
Frequencies and Descriptive Statistics
Effective presentation of study results, in presentation or manuscript form, typically starts with frequencies and descriptive statistics (ie, mean, medians, standard deviations). One can get a better sense of the variables by examining these data to determine whether a balanced and sufficient research design exists. Frequencies also inform on missing data and give a sense of outliers (will be discussed below).
Luckily, software programs are available to conduct exploratory data analysis. For this chapter, we will be examining the following research question.
RQ: Are there differences in drug life (length of effect) for Drug 23 based on the administration site?
A more precise hypothesis could be: Is drug 23 longer-lasting when administered via site A compared to site B?
To address this research question, exploratory data analysis is conducted. First, it is essential to start with the frequencies of the variables. To keep things simple, only variables of minutes (drug life effect) and administration site (A vs B) are included. See Image. Figure 1 for outputs for frequencies.
Figure 1 shows that the administration site appears to be a balanced design with 50 individuals in each group. The excerpt for minutes frequencies is the bottom portion of Figure 1 and shows how many cases fell into each time frame with the cumulative percent on the right-hand side. In examining Figure 1, one suspiciously low measurement (135) was observed, considering time variables. If a data point seems inaccurate, a researcher should find this case and confirm if this was an entry error. For the sake of this review, the authors state that this was an entry error and should have been entered 535 and not 135. Had the analysis occurred without checking this, the data analysis, results, and conclusions would have been invalid. When finding any entry errors and determining how groups are balanced, potential missing data is explored. If not responsibly evaluated, missing values can nullify results.
After replacing the incorrect 135 with 535, descriptive statistics, including the mean, median, mode, minimum/maximum scores, and standard deviation were examined. Output for the research example for the variable of minutes can be seen in Figure 2. Observe each variable to ensure that the mean seems reasonable and that the minimum and maximum are within an appropriate range based on medical competence or an available codebook. One assumption common in statistical analyses is a normal distribution. Image . Figure 2 shows that the mode differs from the mean and the median. We have visualization tools such as histograms to examine these scores for normality and outliers before making decisions.
Histograms are useful in assessing normality, as many statistical tests (eg, ANOVA and regression) assume the data have a normal distribution. When data deviate from a normal distribution, it is quantified using skewness and kurtosis. [1] Skewness occurs when one tail of the curve is longer. If the tail is lengthier on the left side of the curve (more cases on the higher values), this would be negatively skewed, whereas if the tail is longer on the right side, it would be positively skewed. Kurtosis is another facet of normality. Positive kurtosis occurs when the center has many values falling in the middle, whereas negative kurtosis occurs when there are very heavy tails. [2]
Additionally, histograms reveal outliers: data points either entered incorrectly or truly very different from the rest of the sample. When there are outliers, one must determine accuracy based on random chance or the error in the experiment and provide strong justification if the decision is to exclude them. [3] Outliers require attention to ensure the data analysis accurately reflects the majority of the data and is not influenced by extreme values; cleaning these outliers can result in better quality decision-making in clinical practice. [4] A common approach to determining if a variable is approximately normally distributed is converting values to z scores and determining if any scores are less than -3 or greater than 3. For a normal distribution, about 99% of scores should lie within three standard deviations of the mean. [5] Importantly, one should not automatically throw out any values outside of this range but consider it in corroboration with the other factors aforementioned. Outliers are relatively common, so when these are prevalent, one must assess the risks and benefits of exclusion. [6]
Image . Figure 3 provides examples of histograms. In Figure 3A, 2 possible outliers causing kurtosis are observed. If values within 3 standard deviations are used, the result in Figure 3B are observed. This histogram appears much closer to an approximately normal distribution with the kurtosis being treated. Remember, all evidence should be considered before eliminating outliers. When reporting outliers in scientific paper outputs, account for the number of outliers excluded and justify why they were excluded.
Boxplots can examine for outliers, assess the range of data, and show differences among groups. Boxplots provide a visual representation of ranges and medians, illustrating differences amongst groups, and are useful in various outlets, including evidence-based medicine. [7] Boxplots provide a picture of data distribution when there are numerous values, and all values cannot be displayed (ie, a scatterplot). [8] Figure 4 illustrates the differences between drug site administration and the length of drug life from the above example.
Image . Figure 4 shows differences with potential clinical impact. Had any outliers existed (data from the histogram were cleaned), they would appear outside the line endpoint. The red boxes represent the middle 50% of scores. The lines within each red box represent the median number of minutes within each administration site. The horizontal lines at the top and bottom of each line connected to the red box represent the 25th and 75th percentiles. In examining the difference boxplots, an overlap in minutes between 2 administration sites were observed: the approximate top 25 percent from site B had the same time noted as the bottom 25 percent at site A. Site B had a median minute amount under 525, whereas administration site A had a length greater than 550. If there were no differences in adverse reactions at site A, analysis of this figure provides evidence that healthcare providers should administer the drug via site A. Researchers could follow by testing a third administration site, site C. Image . Figure 5 shows what would happen if site C led to a longer drug life compared to site A.
Figure 5 displays the same site A data as Figure 4, but something looks different. The significant variance at site C makes site A’s variance appear smaller. In order words, patients who were administered the drug via site C had a larger range of scores. Thus, some patients experience a longer half-life when the drug is administered via site C than the median of site A; however, the broad range (lack of accuracy) and lower median should be the focus. The precision of minutes is much more compacted in site A. Therefore, the median is higher, and the range is more precise. One may conclude that this makes site A a more desirable site.
- Clinical Significance
Ultimately, by understanding basic exploratory data methods, medical researchers and consumers of research can make quality and data-informed decisions. These data-informed decisions will result in the ability to appraise the clinical significance of research outputs. By overlooking these fundamentals in statistics, critical errors in judgment can occur.
- Nursing, Allied Health, and Interprofessional Team Interventions
All interprofessional healthcare team members need to be at least familiar with, if not well-versed in, these statistical analyses so they can read and interpret study data and apply the data implications in their everyday practice. This approach allows all practitioners to remain abreast of the latest developments and provides valuable data for evidence-based medicine, ultimately leading to improved patient outcomes.
- Review Questions
- Access free multiple choice questions on this topic.
- Comment on this article.
Exploratory Data Analysis Figure 1 Contributed by Martin Huecker, MD and Jacob Shreffler, PhD
Exploratory Data Analysis Figure 2 Contributed by Martin Huecker, MD and Jacob Shreffler, PhD
Exploratory Data Analysis Figure 3 Contributed by Martin Huecker, MD and Jacob Shreffler, PhD
Exploratory Data Analysis Figure 4 Contributed by Martin Huecker, MD and Jacob Shreffler, PhD
Exploratory Data Analysis Figure 5 Contributed by Martin Huecker, MD and Jacob Shreffler, PhD
Disclosure: Jacob Shreffler declares no relevant financial relationships with ineligible companies.
Disclosure: Martin Huecker declares no relevant financial relationships with ineligible companies.
This book is distributed under the terms of the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0) ( http://creativecommons.org/licenses/by-nc-nd/4.0/ ), which permits others to distribute the work, provided that the article is not altered or used commercially. You are not required to obtain permission to distribute this article, provided that you credit the author and journal.
- Cite this Page Shreffler J, Huecker MR. Exploratory Data Analysis: Frequencies, Descriptive Statistics, Histograms, and Boxplots. [Updated 2023 Nov 3]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-.
In this Page
Bulk download.
- Bulk download StatPearls data from FTP
Related information
- PMC PubMed Central citations
- PubMed Links to PubMed
Similar articles in PubMed
- Contour boxplots: a method for characterizing uncertainty in feature sets from simulation ensembles. [IEEE Trans Vis Comput Graph. 2...] Contour boxplots: a method for characterizing uncertainty in feature sets from simulation ensembles. Whitaker RT, Mirzargar M, Kirby RM. IEEE Trans Vis Comput Graph. 2013 Dec; 19(12):2713-22.
- Review Univariate Outliers: A Conceptual Overview for the Nurse Researcher. [Can J Nurs Res. 2019] Review Univariate Outliers: A Conceptual Overview for the Nurse Researcher. Mowbray FI, Fox-Wasylyshyn SM, El-Masri MM. Can J Nurs Res. 2019 Mar; 51(1):31-37. Epub 2018 Jul 3.
- [Descriptive statistics]. [Rev Alerg Mex. 2016] [Descriptive statistics]. Rendón-Macías ME, Villasís-Keever MÁ, Miranda-Novales MG. Rev Alerg Mex. 2016 Oct-Dec; 63(4):397-407.
- An exploratory data analysis of electroencephalograms using the functional boxplots approach. [Front Neurosci. 2015] An exploratory data analysis of electroencephalograms using the functional boxplots approach. Ngo D, Sun Y, Genton MG, Wu J, Srinivasan R, Cramer SC, Ombao H. Front Neurosci. 2015; 9:282. Epub 2015 Aug 19.
- Review Graphics and statistics for cardiology: comparing categorical and continuous variables. [Heart. 2016] Review Graphics and statistics for cardiology: comparing categorical and continuous variables. Rice K, Lumley T. Heart. 2016 Mar; 102(5):349-55. Epub 2016 Jan 27.
Recent Activity
- Exploratory Data Analysis: Frequencies, Descriptive Statistics, Histograms, and ... Exploratory Data Analysis: Frequencies, Descriptive Statistics, Histograms, and Boxplots - StatPearls
Your browsing activity is empty.
Activity recording is turned off.
Turn recording back on
Connect with NLM
National Library of Medicine 8600 Rockville Pike Bethesda, MD 20894
Web Policies FOIA HHS Vulnerability Disclosure
Help Accessibility Careers
Click through the PLOS taxonomy to find articles in your field.
For more information about PLOS Subject Areas, click here .
Loading metrics
Open Access
Peer-reviewed
Research Article
Exploratory data analysis of a clinical study group: Development of a procedure for exploring multidimensional data
Roles Conceptualization, Data curation, Formal analysis, Methodology, Resources, Software, Visualization, Writing – original draft
* E-mail: [email protected] (BMK); [email protected] (LL)
Affiliation Department of Biomedical Engineering, Faculty of Fundamental Problems of Technology, Wroclaw University of Science and Technology, Wroclaw, Poland
Roles Conceptualization, Project administration, Validation, Writing – review & editing
Affiliation Department of Health Promotion, Faculty of Physiotherapy University School of Physical Education, Wroclaw, Poland
Roles Data curation, Investigation
Affiliations Mossakowski Medical Research Centre, Polish Academy of Sciences, Warsaw, Poland, International Institute of Molecular and Cell Biology, Warsaw, Poland
Roles Conceptualization, Project administration, Supervision, Writing – review & editing
Affiliation Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, Wroclaw, Poland
- Bogumil M. Konopka,
- Felicja Lwow,
- Magdalena Owczarz,
- Łukasz Łaczmański

- Published: August 23, 2018
- https://doi.org/10.1371/journal.pone.0201950
- Reader Comments
Thorough knowledge of the structure of analyzed data allows to form detailed scientific hypotheses and research questions. The structure of data can be revealed with methods for exploratory data analysis. Due to multitude of available methods, selecting those which will work together well and facilitate data interpretation is not an easy task. In this work we present a well fitted set of tools for a complete exploratory analysis of a clinical dataset and perform a case study analysis on a set of 515 patients. The proposed procedure comprises several steps: 1) robust data normalization, 2) outlier detection with Mahalanobis (MD) and robust Mahalanobis distances (rMD), 3) hierarchical clustering with Ward’s algorithm, 4) Principal Component Analysis with biplot vectors. The analyzed set comprised elderly patients that participated in the PolSenior project. Each patient was characterized by over 40 biochemical and socio-geographical attributes. Introductory analysis showed that the case-study dataset comprises two clusters separated along the axis of sex hormone attributes. Further analysis was carried out separately for male and female patients. The most optimal partitioning in the male set resulted in five subgroups. Two of them were related to diseased patients: 1) diabetes and 2) hypogonadism patients. Analysis of the female set suggested that it was more homogeneous than the male dataset. No evidence of pathological patient subgroups was found. In the study we showed that outlier detection with MD and rMD allows not only to identify outliers, but can also assess the heterogeneity of a dataset. The case study proved that our procedure is well suited for identification and visualization of biologically meaningful patient subgroups.
Citation: Konopka BM, Lwow F, Owczarz M, Łaczmański Ł (2018) Exploratory data analysis of a clinical study group: Development of a procedure for exploring multidimensional data. PLoS ONE 13(8): e0201950. https://doi.org/10.1371/journal.pone.0201950
Editor: Surinder K. Batra, University of Nebraska Medical Center, UNITED STATES
Received: March 8, 2018; Accepted: July 25, 2018; Published: August 23, 2018
Copyright: © 2018 Konopka et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All relevant data are within the paper and its Supporting Information files.
Funding: The project was partly supported by Wroclaw Centre of Biotechnology through the programme The Leading National Research Centre (KNOW) for years 2014-2018. BMK would like to acknowledge the funding from the statuary fund of the Department of Biomedical Engineering, Wroclaw University of Science and Technology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Introduction
Thorough knowledge of the structure of analyzed data allows to form detailed scientific hypotheses and research questions. It is crucial for correct interpretation of conducted experiments. This is especially important in case of investigations where the researcher does not directly control the conditions or the investigated objects. Clinical or epidemiological studies can be examples of such investigations. Here we will present a case-study analysis of a group of 515 elderly participants of an epidemiological study. Despite the fact that usually participants of clinical studies go through a qualification procedure, fill in detailed question forms and need to meet requirements regarding biochemical parameters, age, health history etc., it may happen that a gathered dataset still contains individuals that should not take part in the study. Their presence in the dataset may significantly influence its final outcome and lead to false conclusions.
The data structure and basic associations between parameters in the data can be revealed with methods for exploratory data analysis, such as clustering or Principal Component Analysis (PCA). Distanced based data analysis methods (including many types of clustering and PCA) are sensitive to data scaling. Therefore data normalization is often needed. Typically this can be performed with Z-score normalization, which assumes normal distribution of values of an attribute. It indicates how many standard deviations an instance of the data is away from the sample mean. Another often used normalization method is the Min-max normalization, which scales an attribute to a 0–1 range. It is especially useful when the bottom and top values of the attribute are limited—for instance due to experimental design. These normalization techniques are sensitive to outliers. The robust Z-score normalization is a modification of the classic Z-score normalization in which median is used instead of the mean and interquartile range is used instead of the standard deviation. These changes minimize the influence of extreme values on the resulting normalization.
Identification of outliers in the data set is another important step in the analysis. Outliers are instances of data that are characterized by extreme attribute values in comparison to the core of the dataset. An outlier can be defined as an instance that was generated by a different process than the rest of instances [ 1 ]. Outliers in single dimensional data can be filtered out with univariate statistic based methods [ 2 ]. However, for high-dimensional data more sophisticated methods need to be used. These methods can be divided into 1) model-based approaches, which assume a model of data—if a data point does not fit the model, it is labelled as an outlier [ 3 ], [ 4 ], 2) proximity-based approaches, which calculate the distance between a data point and all other data—outliers are points that show significantly different distances [ 5 ], [ 6 ] 3) angle-based approaches, which calculate the angles between a data point and all other data, outliers are points that acquire small fluctuations of angles [ 7 ]. Thorough reviews of outlier detection techniques can be found in [ 8 ], [ 9 ] and [ 10 ].
The structure of pre-processed data can be investigated with clustering techniques. These fall into several main categories: 1) hierarchical clustering, 2) partitioning relocation methods (which include various versions of K-means and K-medoids), 3) density-based partitioning, and 4) grid-based partitioning, which performs segmentation of attribute space and agglomeration of similar segments. For a review see [ 11 ]. Among these, hierarchical clustering is associated with probably the clearest way of visualization, i.e. the dendrogram also called the clustering tree, which allows detailed investigation of every clustering step. That is why it is especially useful in data exploration. Clustering quality can be verified quantitatively with clustering validation indices, such as Dunn index [ 12 ], Davies-Bouildin index [ 13 ] or silhouette values [ 14 ].
Data visualization is an extremely important element of data exploration analysis. It allows to connect facts and form conclusions based on the outcome of other steps of the analysis. A classical method for visualization of multidimensional data is PCA [ 15 ], which allows to reduce the number of dimensions needed to depict a dataset without a significant loss of information. However this can also be performed with multidimensional scaling [ 16 ] or some other nonlinear dimensionality reduction techniques [ 17 ].
As it can be seen from this short introduction, when facing the problem of getting to know a new dataset, a researcher has a plethora of exploratory tools to choose from. Selecting methods that will work together and facilitate revealing the structure of the data is not an easy task. In this work we present a well fitted set of tools for a complete exploratory analysis of a clinical study dataset. We perform a case-study analysis in which we address the most important questions that need to be asked prior to most studies: are there any significant outliers in the dataset? What subgroups make up for the dataset? What are the characteristics of particular subgroups? And finally, what are the biological reasons that underlie such dataset structure?
Dataset description
The presented analysis is part of a project which aims at investigating the relation between certain polymorphisms of a gene–Vitamin D Receptor and sex hormone levels in elderly people. The research sample was chosen from the PolSenior study [ 18 ]—a project that aims at investigating the interrelations between health, genetics and social status in advanced age in Polish population.
The dataset consisted of 515 participants– 238 women, and 277 men, whose age was in the range 55–102 years. Each participant was described by 23 numeric and 21 nominal attributes ( S1 Table ). Numeric attributes contain biophysical and biochemical parameters, such as AGE, WEIGHT and BLOOD INSULIN CONCENTRATION. Nominal attributes include socio-geographical data such as COUNTRY REGION, CITY POPULATION, and also SEASON and MONTH. The full list of attributes and their description is given in Tables 1 and 2 . The study was approved by Bioethical Committee of the Medical University of Silesia (KNW-6501-38/I/08) and informed written consent, including consent for genetic studies, was obtained from all of the subjects before testing.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pone.0201950.t001
https://doi.org/10.1371/journal.pone.0201950.t002
Data exploration procedure
As mentioned in the introduction: data visualization and clustering are crucial for understanding the data at hand. These were key elements of the procedure proposed in the study. In order to visualize multidimensional data in a two dimensional space, dimension reduction has to be performed. We used PCA which is a classical method, available in most statistical packages. Using PCA requires data scaling, otherwise attributes with highest variance may dominate the outcome. For the same reason outliers need to be detected and removed.
The exploratory analysis was carried out in two stages. First, we conducted the exploratory analysis based on numeric attributes ( Table 1 ) using the following procedure: 1) normalization, 2) Principal Component Analysis, 3) Outlier detection and removal, 4) clustering. After that, clustering was repeated with the nominal/categorical attributes added ( Table 2 ). We performed the analysis in two stages because processing numerical data is more straightforward–most analysis algorithms were designed to treat numerical data. Processing nominal data requires additional actions to transform from the nominal attribute space to a numerical one and the results need to be analyzed with great caution.

Normalization
Principal Component Analysis (PCA)
Basic R package function prcomp was used for calculation of principal components (PCs). The PC biplot was used for visualization of PCs along with variability and contributions of original attributes [ 21 ]. PCA was carried out on normalized data.
Outlier detection
Two approaches were used to detect outlying samples.
The robust Minimum Covariance Determinant (MCD) is a modification of Mahalanobis distance as defined in [ 3 ]. It is also called the robust Mahalanobis Distance (rMD). The MCD algorithm is an iterative procedure. The steps are:
- Chose a subset H of size h .
- Sort all samples in terms of rMD ( x i ).
- Choose a new subset H 2 of h samples with the smallest rMD .
- Repeat 1–5 untill det ( S k ) = 0 or det ( S k ) = det ( S k − 1 ), where k is the iteration number.
Both MD and rMD were calculated using the ‘chemometrics’ R Package [ 22 ].
Hierarchical clustering analysis
The main clustering approach used was hierarchical clustering. It was performed in two steps. First, samples were clustered based only on numerical attributes. Then, nominal attributes were incorporated for a joined cluster analysis. Nominal attributes were binarized and then rescaled, so that 0 and 1 equaled the I-st and the III-rd quartile of the distribution of all numerical values. This way the center of the data remained unchanged upon addition of nominal attributes. Simultaneously, we performed clustering of attributes. We used hierarchical agglomerative clustering using Ward method, which minimizes the change in variance resulting from fusion of two clusters [ 23 ]. Technically, calculations were carried out with hclust R function with the “ward.D2” method.
Dunn [ 12 ] and Davies–Bouldin [ 13 ] indices were used to support this cluster analysis and index proper number of clusters. The indexes were calculated using the ‘clv’ R Package [ 24 ].
During calculation of Dunn and DB indices we chose diam(c) to be the average distance between cluster members and cluster centroids, and d(c i , c j ) to be the distance between centroids of compared clusters. The choice was implied by the fact that Ward’s clustering algorithm minimizes the within-cluster variance which is defined as the average distance between cluster members and cluster centroids, and also maximizes the inter-cluster variance which is based on centroid locations [ 23 ]. Therefore, such a choice of measures for Dunn and DB gives the best insight into the outcome of clustering.
Additional cluster analysis
Hierarchical clustering analysis of the male set was additionally supported with three other clustering techniques: 1) density-based DBSCAN clustering [ 25 ], 2) clustering based on PCAs and 3) biclustering in order to verify the main conclusions.
Density Based clustering depends on two input parameters, i.e. number of neighbors required to start a new cluster– K , and the distance defining the neighborhood of a point– epsilon . K was set to 3 based on visual inspection of the dataset, while epsilon was set to 4 based on k Nearest Neighbor Distance plot (see Results ). The choice was the y-value beyond which the distances increased rapidly. We used the DBSCAN R package implementation of the algorithm [ 26 ].
PCA-based clustering was performed on top 7 PCs, which accounted for 70% of data variance. The same routine as for main hierarchical clustering was used, i.e. euclidean distance and Wards algorithm as implemented in R stats package.
The biclustering approach used was the Plaid Models clustering [ 27 ], which allows to identify subsets of rows and columns with coherent values. In case of the analyzed dataset those subsets could be regarded as subgroups of patients presenting similar dependence of particular attributes. The biclust package implementation of the algorithm was used [ 28 ].
Statistical testing
Significance of differences between all clusters in terms of particular attributes was first tested with the Kruskal-Wallis test [ 29 ]–h 0 : distributions are the same in all groups. Then paired Wilcoxon rank sum test with Bonferroni correction was used to evaluate the head-to-head difference significance. Both are non-parametric test available in R basic {stats} package.
Results & discussion
Introductory analysis.
Firstly, raw data were normalized using the robust Z-score normalization then PCA was carried out. The plot of first two components shows that there are significant outliers in the data set ( Fig 1A ). The first component clearly dominates the remaining ones ( Fig 1B ). The main contribution to the first component comes from the INSULINE level (data not shown) due to increased variability caused by outliers. MD vs rMD plot shows that the majority of data forms a core ( Fig 1C –grey points) and also confirms the presence of significantly outlying samples ( Fig 1C –red points).
A) PCA carried out on full dataset. B) standard deviations of first 10 PCs indicate that the first PC dominates the variability of the dataset. C) The MD vs rMD plot allows to identify the most distant outliers (red points). D) PCA carried out after removal of most distant samples shows that male and female patients form two distinct clusters.
https://doi.org/10.1371/journal.pone.0201950.g001
In order to get an overall look at the core of data we used arbitrarily set MD and rMD thresholds to remove the most distant outliers, 6.5 and 15 respectively ( Fig 1C –dashed lines). The thresholds were selected so that only the core of the data remained.
The plot of two first components, calculated after removing outlying points, reveals that samples are grouped in two clusters, consisting of male and female patients respectively ( Fig 1D ). The biplot [ 1 ] allows to visualize contributions of original attributes to particular PCs in the form of vectors. For instance if a patient had a level of ESTRADIOL higher than average, then in the PCA with biplot vectors he/she would be moved away from the center of the plot in the direction pointed by the ESTRADIOL vector. It can be seen that the two acquired clusters are separated along an axis formed by attributes such as: ESTRADIOL, TESTOSTERONE, FEI, FAI, FSH, which are sex hormones ( Fig 1D –red vectors). Such strong separation suggests that further analysis should be carried out separately for male and female patients. The position of particular samples in Fig 1D is also strongly influenced by a group of attributes perpendicular to the sex hormone axis. These attributes are generally related to metabolism: such as GLUCOSE, INSULINE, FAT, WEIGHT etc. The fact that these attributes are perpendicular to the sex hormone axis suggested they are unrelated to patient sex.
Male set analysis
In the first part of male set analysis all 277 male patients with all 23 numeric attributes from the raw dataset were analyzed. Again robust Z-score normalization was performed.
According to MD there are 22 outliers in the dataset. These points clearly stand out in terms of MD values from the rest of the set ( Fig 2A –red points). In terms of rMD there are many more candidate outliers, i.e. 124 samples. Both measures are consistent with regard to MD outliers—all samples pointed as outliers by the classic MD were also outliers in terms of rMD, what is more these were among the points with the highest rMD values ( Fig 2b –red points). The fact that rMD indicated almost half of the dataset as outliers may suggest that the set is heterogeneous.
A) according to classic MD, B) according to rMD. Outliers according to MD are colored red in both plots. The dashed line denotes the 0.99 quantile threshold for Chi2 distribution used for flagging outliers.
https://doi.org/10.1371/journal.pone.0201950.g002
The MD vs rMD plot reveals that the data can be divided into three groups: 1) 155 samples that form the core of the set ( Fig 3 –gray points), 2) 100 samples that are rMD outliers only ( Fig 3 –blue points) 3) 22 samples that are outliers according to both MD and rMD ( Fig 3 red points marked blue). This shows that the classic MD is more conservative in terms marking outliers than the rMD. Both measures MD and rMD calculate the distance of data points from the data center. However while MD uses all points to determine the data center location, rMD uses only a subset of points that are the closest to the center (see Methods for more details). If a dataset consists of two subsets of points then rMD may use only one of them two determine the center of the data (this depends on the sizes of subsets). In such a situation points from the other set may be seen as outliers in terms of rMD. That is why this measure can be successfully used to state whether the set is homo- or heterogeneous.
Outliers were marked with blue and red points for rMD and MD respectively. All MD outliers are also rMD outliers.
https://doi.org/10.1371/journal.pone.0201950.g003
Hierarchical clustering
We performed two rounds of clustering: 1) clustering of attributes–attributes were treated as instances and patients were treated as attributes, 2) clustering of patients—patients were treated as instances and their parameters were treated as attributes.
Clustering of attributes showed that there are three main groups of parameters ( Fig 4A —top panel), i.e. age-related parameters (FSH, SHGB, ICTP, AGE, OPG), cholesterol and sex-hormone related parameters (including TESTOSTERONE, ESTRADIOL, DHEA), and metabolism related parameters (such as FAT, WEIGHT, BMI, GLUCOSE and INSULINE). This division was also confirmed in the PCA biplot, which depicts three groups of attribute vectors pointing in similar directions ( Fig 5A ). These three groups correspond well to groups revealed by clustering.
A) top panel–attribute clustering tree, left panel–patient clustering tree, central panel–dataset heatmap; branch length is proportional to distances between clusters B) Davies Bouldin index for patient partitioning into 2–10 clusters C) Dunn index for patient partitioning into 2–10 clusters.
https://doi.org/10.1371/journal.pone.0201950.g004
A) in PCA biplot, B) MD vs rMD metrics.
https://doi.org/10.1371/journal.pone.0201950.g005
The patient clustering tree is presented in Fig 4A –left panel. Acquired partitioning was validated using Davies-Buildin (DB) and Dunn indices at different tree cut levels, i.e divisions into 2 to 10 clusters were analyzed. Neither DB nor Dunn index clearly indicated which cluster partitioning is the most appropriate ( Fig 4B and 4C ). In case of the DB good partitioning is indicated by small values. As depicted in Fig 4B , DB index decreases as the number of clusters increases, with a local minimum formed for the division in to 5 groups. In case of the Dunn index a good partitioning is indicated by high values. The highest values can be observed for partitioning into 2 and 3 clusters. However, a local maximum can be observed at the division into 5 groups ( Fig 4C ). Since both indices emphasized clustering into 5 groups, this partitioning is analyzed in greater details.
Partitioning the set into 5 groups results in two large clusters- cl #1 and cl #5, of 89 and 80 samples respectively and three smaller clusters cl #2–24 samples, cl #3–24 samples and cl #4–38 samples. According to MD and rMD metrics clusters #1, #2 and #5 form the core of the data as shown in Fig 5B , while clusters #3 and #4 deviate from the core and form the majority of RD outliers ( Fig 5B ).
The significance of differences between all clusters in terms of particular attributes were tested first with the Kruskal-Wallis test [ 29 ] and then paired Wilcoxon rank sum test with Bonferroni correction. In Fig 6 p-values of all-vs-all Wilcoxon tests were shown.
Values in red denote p-values.
https://doi.org/10.1371/journal.pone.0201950.g006
Cluster #3 is characterized by significantly elevated levels of INSULINE and GLUCOSE. This is clearly visible in the clustering heatmap as a bright area in INS and GLUCOSE columns ( Fig 4A ). In PCA bioplot members of the cluster are localized far away from the center of the dataset along INS and GLUCOSE vectors ( Fig 5A ). The significance of difference between #3 and members of other clusters was confirmed by statistical tests ( Fig 6 ). We suspect this cluster may be a group of putative diabetes patients. Cluster #4 is characterized by exceptionally high levels of FSH and ICTP hormones, which are accompanied by low level TESTOSTERONE and decreased ESTRADIOL. The group is also characterized by greater AGE values. FAI and FEI attributes are also low in this group of patients, however this was expected since TESTOSTERON and FAI as well as ESTRADIOL and FEI are related attributes. In the PCA biplot ( Fig 5A ) Members of cluster #4 are localized far away from the center of the dataset along the FSH and ICTP vectors. High FSH and low serum level of TESTOSTERONE may indicate that these patients suffer from primary hypogonadism [ 30 ].
The core of the data in terms of MD and rMD is formed by clusters #1, #2 and #5. Cluster #2 is the smallest of them. As featured by the dendrogram ( Fig 4A –left panel) it is closely related to cluster #5. With the main difference between them being the elevated levels of cholesterol (CHOL.LDL, CHOL.HDL, and CHOL.TOTAL). Members of both clusters are characterized by relatively high TESTOSTERONE levels.
The largest clusters #1 and #5 are hard to be characterized since they form a reference point for describing remaining clusters. The main difference between them comes from metabolism-related attributes: WEIGHT, WAISTLINE, BMI, HIP.GIRTH, FAT, TGC, INS, GLUCOSE. This can be observed in the clustering heat map as a darker patch in the region of cluster #5 ( Fig 4A ). The difference became more evident after addition of categorical data, which included metabolic phenotype classifications (see next section). The clusters also differ in terms of SHGB and FEI, FAI levels. In the PCA biplot members of cluster #5 are shifted in the opposite direction to the one pointed by metabolic attributes ( Fig 5A ) and also towards the SHGB direction. The latter confirms higher SGHB values in this cluster. Quite interestingly members of both largest clusters can be found not only in the core of the data but also in the rMD outlier group ( Fig 5B ), which means that further division might reveal some interpretable subgroups.
Addition of categorical data
Categorical attributes were transformed to binary attributes and scaled as described in Methods section. Hierarchical clustering with Wards algorithm was repeated. Clustering validation Davies-Bouldin and Dunn indexes both indicated division into three clusters as the most appropriate partitioning (data not shown). Two of the clusters could be easily identified as outlier clusters #3 (aberrant GLUCOSE and INS levels) and #4 (aberrant FSH and ICTP) from the numerical attribute clustering analysis. The third cluster forms the core of the data which includes clusters #1, #2 and #5 ( Fig 7 —left panel). Obesity phenotype attributes present in in the set of categorical attributes confirmed that the main difference between cluster #1 and clusters #2 and #5 is related to metabolism–dark patch in OBESITY_PHENOOZZM and OBESTITY_PHENO_FLOMWD and light patch in OBESITY_PHENO_FLMONW ( Fig 7 —heatmap).
https://doi.org/10.1371/journal.pone.0201950.g007
Other clustering approaches
Applying additional methodologically divergent approaches may strengthen the final conclusions or suggest other optional viewpoints. We supported the main clustering analysis with three alternative approaches: density-based DBSCAN clustering, hierarchical clustering based on top principal components and biclustering focused on identification of coherent values. While PC based clustering and biclustering approaches led to conclusions compliant with those already presented, the density based approach was unable to uncover the underlying structure of the data. The majority of samples fell into a single cluster and only a few marginal samples were marked as noise (see S1B Fig ). Most probably this is due to the fact that the subgroups overlap and also are characterized by similar point densities, which make them hard to separate by the DBSCAN algorithm. However, the method was successfully applied to support outlier detection. When we ran the algorithm on the dataset containing outliers, the algorithm marked 31 samples as noise. All of them were also marked as outliers by either MD or RD distances ( S1 Table ).
Opposite to DBSCAN clustering–the clustering based on top 7 PCs, which accounted for 70% of data variance, resulted in a partitioning very similar to the one acquired by the main clustering approach ( S2 Fig ).
Finally, the main conclusions were also supported by the outcome of the biclustering plaid model analysis. All significant clusters and relations were found. However, the clusters were smaller and the outcomes were subject to some the randomness due to the nature of the clustering algorithm ( S3A–S3D Fig ).
Female set analysis
The female set was analyzed using the same methodology that was applied in male set analysis. The set included 238 patients with 23 numeric attributes. Data were normalized with the Z-score robust normalization, then outlier analysis was carried out with MD and robust MD distances, finally we performed hierarchical clustering analysis supported with DB and Dunn clustering validation indices.
Outlier analysis in the female set indicates 70 and 20 robust MD and MD outliers respectively. All MD outliers were also robust MD outliers. The robust MD vs MD plot differs significantly from the plot acquired in the male set analysis–points are more condensed and cannot easily divided into subgroups ( Fig 8 ). Although there are many outliers according to rMD, it seems that only a few of them are actual outliers. The majority of rMD outliers remain quite close to the core of the dataset in terms of MD. This suggests that female dataset is more homogeneous than the male dataset.
MD and rMD are consistent–most points lie on a straight line.
https://doi.org/10.1371/journal.pone.0201950.g008
Hierarchical clustering of attributes confirmed the division revealed in male set analysis, i.e. three attribute groups were identified: age-related parameters (FSH, SHGB, ICTP, AGE, OPG), cholesterol and sex-hormone related parameters (including TESTOSTERONE, ESTRADIOL, DHEA), and metabolism related parameters ( Fig 9A –top panel). The HDL Cholesterol level was an exception–in this analysis it is part of the age related attribute group.
(A) The violet line and labels on the dendrogram denote the best partitioning according to cluster validation indices. Davies- Bouldin index (B) and Dunn index(C) indicate that partitioning the set into 2 or 3 clusters are the best choices for further analysis (low DB and high Dunn values).
https://doi.org/10.1371/journal.pone.0201950.g009
According to DB and Dunn indices the optimal division of female patients includes two or three groups ( Fig 9B and 9C ). We analyzed the three cluster division as it is more informative. In this case cluster #1 consists of 71 patients. These patients are characterized by low values of metabolic parameters ( Fig 9A –heatmap, dark path in GLUCOSE, INS, FAT and others), and elevated levels of SHGB, FSH, CHOL.HDL. Cluster #2 groups 131 patients. It forms the core of the dataset and probably represents the majority of population. Finally cluster #3, a cluster of 14 patients with high levels of metabolic parameters (GLUCOSE, INS, FAT and others) but also elevated levels of TESTOSTERONE and ESTRADIOL.
The biplot visualization of the data is consistent with both: clustering of attributes and clustering of patients. The contributions of particular attributes in PCs confirm the relations between parameters–metabolic parameters and hormone related parameters form two well distinguishable groups of similarly pointing vectors. The third group is more diverse, but the sub groups are correct, i.e OPG, AGE and ICTP form one group and FSH SHGB and CHOL.HDL form a second group of vectors ( Fig 9 red arrows). The distribution of patients in the biplot is also consistent with the clustering. Members of cluster #1 are localized in the region pointed by SHGB, FSH and CHOL.HDL vectors, and opposite the direction of metabolic attributes. Members of cluster #2 are in the center of the plot, while members of cluster #3 are shifted away from the origin mainly in the direction of metabolic attributes.
Over all the PCA plot of the female set is more homogeneous in comparison to the PCA in the male set analysis ( Fig 10 ). Samples present are more evenly distributed around the origin, while in the male set subgroups could be easily distinguished. This suggests that in the female set there are no pathological groups of patients that could be recognized based on the set of attributes at hand. However still, there are some patients that should be investigated and verified prior to including them in further studies (for instance three patients in cluster #3 furthest away from the origin).
Patients form a quite condensed cloud of point (we just a few exceptions). The clusters result from natural biological variation rather than from pathologies.
https://doi.org/10.1371/journal.pone.0201950.g010
In this work we presented a data exploratory analysis of a clinical study group. Each patient was described by over 40 numerical and nominal attributes. The aim of the study was to reveal the structure of the data, i.e. verify whether the population of patients is homogenous or whether subpopulations are present. We also wanted to characterize identified subgroups and to investigate basic relations between attributes. The analysis was performed with a set of methods that were specially selected to work well together. First a robust normalization technique was used. Then MD based outlier detection methods, hierarchical clustering with Wards algorithm and PCA visualization was performed. Since all these methods take in to account the correlation and variance of data attributes, their outcomes were consistent. We have shown that the MD/rMD analysis allows not only to identify outliers but can also be used to assess the heterogeneity of a dataset. PCA together with the biplot allowed to characterize data instances and explain the acquired clustering. The analysis was additionally supported by three alternative clustering approaches, which strengthen the main conclusions and contributed to better understanding of the data.
Several important biological conclusion can be drawn. The study showed significant differences between male and female patients. In the male set we managed to identify five distinct patient groups, two of which were recognized as clusters of putatively diseased patients. In further analysis this structure should be taken into account. One should consider testing scientific hypothesis separately in each of identified subgroups. Depending on the aims of subsequent investigation some of the groups should be removed or treated in a special way.
The female set was more homogenous in comparison to the male set and the clusters we identified were not recognized as pathological. However, still one might also consider performing further investigations separately in the identified subgroups.
Neglecting the fact of existence of patient subgroups might make it impossible to reveal important biological phenomena or in the worst case lead to false conclusions.
Supporting information
S1 table. analysis data..
https://doi.org/10.1371/journal.pone.0201950.s001
S2 Table. Comparison of sample labeling by density-based clustering and Mahalanobis Distances.
https://doi.org/10.1371/journal.pone.0201950.s002
S1 Code. Exploratory analysis code.
https://doi.org/10.1371/journal.pone.0201950.s003
S1 Fig. Density based clustering on the male dataset.
A) the parameters chosen for clustering were K = 3 neighbors and epsilon = 4 (based on the elbow method), B) density clustering failed to confirm the structure of the data revealed by hierarchical clustering by managed to mark marginal points (zero’s) and could be used for outlier detection.
https://doi.org/10.1371/journal.pone.0201950.s004
S2 Fig. Hierarchical clustering based on first 7 Principal Components shows high accordance with clustering based on full attribute set.
Most importantly clusters of patients with high levels of FSH or GLUCOSE/INSULIN were found (blue and green cluster respectively).
https://doi.org/10.1371/journal.pone.0201950.s005
S3 Fig. Subclusters identified by plaid model biclustering.
The analysis resulted in identifying the two important outlier clusters: A) the cluster with elevated INSULIN and GLUCOSE levels and B) patients with elevated FSH levels. In addition two other patient subgroups were found: C) one showing a dependence of hormone and cholesterol related attributes and D) group of patients with simultaneously elevated SHGB and CHOL.HDL levels.
https://doi.org/10.1371/journal.pone.0201950.s006
Acknowledgments
Data analyzed in the case-study were gathered in the PolSenior study. We thank all people engaged in the project. In particular we would like to thank prof. Andrzej Milewicz, prof. Malgorzata Mossakowska, prof. Monika Puzianowska-Kuznicka, prof. Ewa Bar-Andziak, prof. Jerzy Chudek.
We would like to thank Dr. Jean-Christophe Nebel for his valuable comments and discussion during preparation of the manuscript.
- 1. Hawkins DM, Identification of Outliers , Chapman and Hall, London–New York 1980
- View Article
- Google Scholar
- 6. Ramaswamy S, Rastogi R, Shim K, Efficient algorithms for mining outliers from large data sets, Proceedings of the 2000 ACM SIGMOD international conference on Management of data 2000; pp. 427–438, 10.1145/335191.335437
- 7. Kriegel HP, Hubert MS, Zimek A, Angle-based outlier detection in high-dimensional data, Proceedings of the 14th ACM SIGKDD international conference on Knowledge discovery and data mining 2008; pp. 444–452
- 8. Ben-Gal I, Outlier Detection , Data Mining and Knowledge Discovery Handbook, 2005 pp 131–146
- 10. Zhang Ji, Advancements of Outlier Detection: A Survey, Transactions on Scalable Information Systems 2013; 13:01–03
- 11. Berkhin P, Survey of clustering data mining techniques. Technical report , Accrue Software , 2002; San Jose, CA
- 15. Jolliffe IT, Principal Component Analysis, Springer Series in Statistics, 2nd ed., Springer, NY, 2002, https://doi.org/10.1002/0470013192.bsa501
- 16. Torgerson WS, Theory & Methods of Scaling , Wiley, NY 1958
- PubMed/NCBI
- 20. Lwow F, Wpływ standaryzowanego wysiłku fizycznego na stres oksydacyjny w aspekcie fenotypu otyłości i polimorfizmu genu receptora beta3-adrenergicznego u kobiet pomenopauzalnych, Wydawnictwo AWF Wrocław, 2010
- 22. Filzmoser P, Varmuza K, Chemometrics R package, https://cran.r-project.org/web/packages/chemometrics/index.html
- 24. Nieweglowski L, clv–Cluster Validation Techniques R package, ( https://cran.r-project.org/web/packages/clv/index.html )
- 25. Ester M, Kriegel HP, Sander J, Xu X, A density-based algorithm for discovering clusters in large spatial databases with noise, Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD-96).
- 26. Hahsler M, DBSCAN R package, ( https://cran.r-project.org/web/packages/dbscan/index.html )
- 28. Kaiser S, Santamaria R, Khamiakova T, Sill M, Theron R, Quintales L, et al. Biclust R package, ( https://cran.r-project.org/web/packages/biclust/index.html )
IEEE Account
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
Welcome to our newly formatted notes. Update you bookmarks accordingly.
2 Exploratory Data Analysis (EDA)
2.1 overview.
Exploratory Data Analysis (EDA) may also be described as data-driven hypothesis generation . Given a complex set of observations, often EDA provides the initial pointers towards various learning techniques. The data is examined for structures that may indicate deeper relationships among cases or variables.
In this lesson, we will focus on both aspects of EDA:
- Numerical summarization
- Data Visualization
This course is based on R software. There are several attractive features of R that make it a software of choice both in academia as well as in industry.
- R is an open-source software and is free to download.
- R is supported by 3,000+ packages to deal with large volumes of data in a wide variety of applications. For instance, the svd() function performs the singular value decomposition in a single line of coding, which cannot be so easily implemented in C, Java or Python.
- R is quite versatile. After an algorithm is developed in R, the program may be sped up by transforming the R codes into other languages.
- R is a mainstream analytical tool.
Reference: * *
- The Popularity of Data Analysis Software by R.A. Muenchen,
- R You Ready for R? by Ashlee Vance
- R Programming for Data Science by Roger Peng
The following diagram shows that in recent times R is gaining popularity as monthly programming discussion traffic shows explosive growth of discussions regarding R.
R has a vibrant user community. As a result of that R has the most website links that point to it.
R can be installed from the CRAN website R-Project following the instructions. Downloading R-Studio is strongly recommended. To develop familiarity with R it is suggested to follow through the material in Introduction to R . For further information refer to the Course Syllabus. Other useful websites on R are Stack Overflow R Questions and R Seek .
One of the objectives of this course is to strengthen the basics in R. The R-Labs given in the textbook are followed closely. Along with the material in the text, two other features in R are introduced.
- R Markdown : This allows the users to knit the R codes and outputs directly into the document.
- R library ggplot2`: A very useful and sophisticated set of plotting functions to produce high-quality graphs
Upon successful completion of this lesson, you should be able to:
- Develop familiarity with R software.
- Application of numerical and visual summarization of data.
- Illustration of the importance of EDA before embarking on sophisticated model building.
2.2 What is Data
Introduction.
Anything that is observed or conceptualized falls under the purview of data. In a somewhat restricted view, data is something that can be measured. Data represent facts or something that has actually taken place, observed and measured. Data may come out of passive observation or active collection. Each data point must be rooted in a physical, demographical or behavioral phenomenon must be unambiguous and measurable. Data is observed in each unit under study and stored in an electronic device.
Definition 2.1 (Data) denotes a collection of objects and their attributes
Definition 2.2 (Attribute) (feature, variable, or field) is a property or characteristic of an object
Definition 2.3 (Collection of Attributes) describe an object (individual, entity, case, or record)
| ID | Sex | Education | Income |
|---|---|---|---|
| 248 | Male | High School | $100,000 |
| 249 | Female | High School | $12,000 |
| 250 | Male | College | $23,000 |
| 251 | Male | Child | $0 |
| 252 | Female | High School | $19,798 |
| 253 | Male | High School | $40,100 |
| 254 | Male | Less than 1st Grade | $2691 |
| 255 | Male | Child | $0 |
| 256 | Male | 11th Grade | $30,000 |
| 257 | Male | Ph.D. | $30686 |
Each Row is an Object and each Column is an Attribute
Often these attributes are referred to as variables. Attributes contain information regarding each unit of observation. Depending on how many different types of information are collected from each unit, the data may be univariate, bivariate or multivariate.
Data can have varied forms and structures but in one criterion they are all the same – data contains information and characteristics that separate one unit or observation from the others.
Types of Attributes
Definition 2.4 (Nominal) Qualitative variables that do not have a natural order, e.g. Hair color, Religion, Residence zipcode of a student
Definition 2.5 (Ordinal) Qualitative variables that have a natural order, e.g. Grades, Rating of a service rendered on a scale of 1-5 (1 is terrible and 5 is excellent), Street numbers in New York City
Definition 2.6 (Interval) Measurements where the difference between two values is meaningful, e.g. Calendar dates, Temperature in Celsius or Fahrenheit
Definition 2.7 (Ratio) Measurements where both difference and ratio are meaningful, e.g. Temperature in Kelvin, Length, Counts
Discrete and Continuous Attributes
Definition 2.8 (Discrete Attribute) A variable or attribute is discrete if it can take a finite or a countably infinite set of values. A discrete variable is often represented as an integer-valued variable. A binary variable is a special case where the attribute can assume only two values, usually represented by 0 and 1. Examples of a discrete variable are the number of birds in a flock; the number of heads realized when a coin is flipped 10 times, etc.
Definition 2.9 (Continuous Attribute) A variable or attribute is continuous if it can take any value in a given range; possibly the range being infinite. Examples of continuous variables are weights and heights of birds, the temperature of a day, etc.
In the hierarchy of data, nominal is at the lowermost rank as it carries the least information. The highest type of data is ratio since it contains the maximum possible information. While analyzing the data, it has to be noted that procedures applicable to a lower data type can be applied for a higher one, but the reverse is not true. Analysis procedure for nominal data can be applied to interval type data, but it is not recommended since such a procedure completely ignores the amount of information an interval type data carries. But the procedures developed for interval or even ratio type data cannot be applied to nominal nor to ordinal data. A prudent analyst should recognize each data type and then decide on the methods applicable.
2.3 Numerical Summarization
Summary statistics.
The vast amount of numbers on a large number of variables need to be properly organized to extract information from them. Broadly speaking there are two methods to summarize data: visual summarization and numerical summarization. Both have their advantages and disadvantages and applied jointly they will get the maximum information from raw data.
Summary statistics are numbers computed from the sample that present a summary of the attributes.
Measures of Location
They are single numbers representing a set of observations. Measures of location also include measures of central tendency. Measures of central tendency can also be taken as the most representative values of the set of observations. The most common measures of location are the Mean, the Median, the Mode, and the Quartiles.
Definition 2.10 (Mean) the arithmetic average of all the observations. The mean equals the sum of all observations divided by the sample size
Definition 2.11 (Median) the middle-most value of the ranked set of observations so that half the observations are greater than the median and the other half is less. Median is a robust measure of central tendency
the most frequently occurring value in the data set. This makes more sense when attributes are not continuous
2.3.2 Quartiles
division points which split data into four equal parts after rank-ordering them.
Division points are called Q1 (the first quartile), Q2 (the second quartile or median), and Q3 (the third quartile)
Note! They are not necessarily four equidistance point on the range of the sample
Similarly, Deciles and Percentiles are defined as division points that divide the rank-ordered data into 10 and 100 equal segments.
Note! that the mean is very sensitive to outliers (extreme or unusual observations) whereas the median is not. The mean is affected if even a single observation is changed. The median, on the other hand, has a 50% breakdown which means that unless 50% values in a sample change, the median will not change.
Measures of Spread
Measures of location are not enough to capture all aspects of the attributes. Measures of dispersion are necessary to understand the variability of the data. The most common measure of dispersion is the Variance, the Standard Deviation, the Interquartile Range and Range.
Definition 2.12 (Variance) measures how far data values lie from the mean . It is defined as the average of the squared differences between the mean and the individual data values
Definition 2.13 (Standard Deviation) is the square root of the variance. It is defined as the average distance between the mean and the individual data values
Definition 2.14 (Interquartile range (IQR)) is the difference between Q3 and Q1. IQR contains the middle 50% of data
Definition 2.15 (Range) is the difference between the maximum and minimum values in the sample
Measures of Skewness
In addition to the measures of location and dispersion, the arrangement of data or the shape of the data distribution is also of considerable interest. The most ‘well-behaved’ distribution is a symmetric distribution where the mean and the median are coincident. The symmetry is lost if there exists a tail in either direction. Skewness measures whether or not a distribution has a single long tail.
Skewness is measured as: \[ \dfrac{\sqrt{n} \left( \Sigma \left(x_{i} - \bar{x} \right)^{3} \right)}{\left(\Sigma \left(x_{i} - \bar{x} \right)^{2}\right)^{\frac{3}{2}}}\]
The figure below gives examples of symmetric and skewed distributions. Note that these diagrams are generated from theoretical distributions and in practice one is likely to see only approximations.
Calculate the answers to these questions then click the icon on the left to reveal the answer.
Suppose we have the data: 3, 5, 6, 9, 0, 10, 1, 3, 7, 4, 8. Calculate the following summary statistics:
- Variance and Standard Deviation
- Mean: (3+5+6+9+0+10+1+3+7+4+8)/11= 5.091.
- Median: The ordered data is 0, 1, 3, 3, 4, 5, 6, 7, 8, 9, 10. Thus, 5 is the median.
- Q1 and Q3: Q1 is 3 and Q3 is 8.
- Variance and Standard Deviation: Variance is 10.491 (=((3-5.091)2+…+(8-5.091)2)/10). Thus, the standard deviation is the square root of 10.491, i.e. 3.239.
- IQR: Q3-Q1=8-3=5.
- Range: max-min=10-0=10.
- Skewness: -0.03.
Measures of Correlation
All the above summary statistics are applicable only for univariate data where information on a single attribute is of interest. Correlation describes the degree of the linear relationship between two attributes, X and Y .
With X taking the values x (1), … , x ( n ) and Y taking the values y (1), … , y ( n ), the sample correlation coefficient is defined as: \[\rho (X,Y)=\dfrac{\sum_{i=1}^{n}\left ( x(i)-\bar{x} \right )\left ( y(i)-\bar{y} \right )}{\left( \sum_{i=1}^{n}\left ( x(i)-\bar{x} \right )^2\sum_{i=1}^{n}\left ( y(i)-\bar{y} \right )^2\right)^\frac{1}{2}}\]
The correlation coefficient is always between -1 (perfect negative linear relationship) and +1 (perfect positive linear relationship). If the correlation coefficient is 0, then there is no linear relationship between X and Y.
In the figure below a set of representative plots are shown for various values of the population correlation coefficient ρ ranging from - 1 to + 1. At the two extreme values, the relation is a perfectly straight line. As the value of ρ approaches 0, the elliptical shape becomes round and then it moves again towards an elliptical shape with the principal axis in the opposite direction.

Try the applet “CorrelationPicture” and “CorrelationPoints” from the University of Colorado at Boulder .
Try the applet “Guess the Correlation” from the Rossman/Chance Applet Collection .
2.3.3 Measures of Similarity and Dissimilarity
Similarity and dissimilarity.
Distance or similarity measures are essential in solving many pattern recognition problems such as classification and clustering. Various distance/similarity measures are available in the literature to compare two data distributions. As the names suggest, a similarity measures how close two distributions are. For multivariate data complex summary methods are developed to answer this question.
Definition 2.16 (Similarity Measure) Numerical measure of how alike two data objects often fall between 0 (no similarity) and 1 (complete similarity)
Definition 2.17 (Dissimilarity Measure) Numerical measure of how different two data objects are range from 0 (objects are alike) to \(\infty\) (objects are different)
Definition 2.18 (Proximity) refers to a similarity or dissimilarity
Similarity/Dissimilarity for Simple Attributes
Here, p and q are the attribute values for two data objects.
| Nominal | \(s=\begin{cases} 1 & \text{ if } p=q \\ 0 & \text{ if } p\neq q \end{cases}\) | \(d=\begin{cases} 0 & \text{ if } p=q \\ 1 & \text{ if } p\neq q \end{cases}\) |
| Ordinal | \(s=1-\dfrac{\left | p-q \right |}{n-1}\) (values mapped to integer 0 to n-1, where n is the number of values) | \(d=\dfrac{\left | p-q \right |}{n-1}\) |
| Interval or Ratio | \(s=1-\left | p-q \right |, s=\frac{1}{1+\left | p-q \right |}\) | \(d=\left | p-q \right |\) |
Distance , such as the Euclidean distance, is a dissimilarity measure and has some well-known properties: Common Properties of Dissimilarity Measures
- d ( p , q ) ≥ 0 for all p and q , and d ( p , q ) = 0 if and only if p = q ,
- d ( p , q ) = d(q,p) for all p and q ,
- d ( p , r ) ≤ d ( p , q ) + d ( q , r ) for all p , q , and r, where d ( p , q ) is the distance (dissimilarity) between points (data objects), p and q .
A distance that satisfies these properties is called a metric . Following is a list of several common distance measures to compare multivariate data. We will assume that the attributes are all continuous.
Euclidean Distance
Assume that we have measurements \(x_{ik}\) , \(i = 1 , \ldots , N\) , on variables \(k = 1 , \dots , p\) (also called attributes).
The Euclidean distance between the i th and j th objects is \[d_E(i, j)=\left(\sum_{k=1}^{p}\left(x_{ik}-x_{jk} \right) ^2\right)^\frac{1}{2}\]
for every pair (i, j) of observations.
The weighted Euclidean distance is: \[d_{WE}(i, j)=\left(\sum_{k=1}^{p}W_k\left(x_{ik}-x_{jk} \right) ^2\right)^\frac{1}{2}\]
If scales of the attributes differ substantially, standardization is necessary.
Minkowski Distance
The Minkowski distance is a generalization of the Euclidean distance.
With the measurement, \(x _ { i k } , i = 1 , \dots , N , k = 1 , \dots , p\) , the Minkowski distance is \[d_M(i, j)=\left(\sum_{k=1}^{p}\left | x_{ik}-x_{jk} \right | ^ \lambda \right)^\frac{1}{\lambda} \]
where \(\lambda \geq 1\) . It is also called the \(L_λ\) metric.
- \(\lambda = 1 : L _ { 1 }\) metric, Manhattan or City-block distance.
- \(\lambda = 2 : L _ { 2 }\) metric, Euclidean distance.
- \(\lambda \rightarrow \infty : L _ { \infty }\) metric, Supremum distance. \[ \lim{\lambda \to \infty}=\left( \sum_{k=1}^{p}\left | x_{ik}-x_{jk} \right | ^ \lambda \right) ^\frac{1}{\lambda} =\text{max}\left( \left | x_{i1}-x_{j1}\right| , ... , \left | x_{ip}-x_{jp}\right| \right) \]
Note that λ and p are two different parameters. Dimension of the data matrix remains finite.
Mahalanobis Distance
Let X be a N × p matrix. Then the \(i^{th}\) row of X is \[x_{i}^{T}=\left( x_{i1}, ... , x_{ip} \right)\]
The Mahalanobis distance is \[d_{MH}(i, j)=\left( \left( x_i - x_j\right)^T \Sigma^{-1} \left( x_i - x_j\right)\right)^\frac{1}{2}\]
where \(∑\) is the p×p sample covariance matrix.
Calculate the answers to these questions by yourself and then click the icon on the left to reveal the answer.
- Calculate the Euclidan distances.
- Calculate the Minkowski distances ( \(\lambda = 1\text{ and }\lambda\rightarrow\infty\) cases).
- Euclidean distances are: \[d _ { E } ( 1,2 ) = \left( ( 1 - 1 ) ^ { 2 } + ( 3 - 2 ) ^ { 2 } + ( 1 - 1 ) ^ { 2 } + ( 2 - 2 ) ^ { 2 } + ( 4 - 1 ) ^ { 2 } \right) ^ { 1 / 2 } = 3.162\]
\[d_{ E } ( 1,3 ) = \left( ( 1 - 2 ) ^ { 2 } + ( 3 - 2 ) ^ { 2 } + ( 1 - 2 ) ^ { 2 } + ( 2 - 2 ) ^ { 2 } + ( 4 - 2 ) ^ { 2 } \right) ^ { 1 / 2 } = 2.646\]
\[d_{ E } ( 2,3 ) = \left( ( 1 - 2 ) ^ { 2 } + ( 2 - 2 ) ^ { 2 } + ( 1 - 2 ) ^ { 2 } + ( 2 - 2 ) ^ { 2 } + ( 1 - 2 ) ^ { 2 } \right) ^ { 1 / 2 } = 1.732\]
- Minkowski distances (when \(\lambda = 1\) ) are:
\[d_ { M } ( 1,2 ) = | 1 - 1 | + | 3 - 2 | + | 1 - 1 | + | 2 - 2 | + | 4 - 1 | = 4\]
\[d_ { M } ( 1,3 ) = | 1 - 2 | + | 3 - 2 | + | 1 - 2 | + | 2 - 2 | + | 4 - 2 | = 5\]
\[d_ { M } ( 2,3 ) = | 1 - 2 | + | 2 - 2 | + | 1 - 2 | + | 2 - 2 | + | 1 - 2 | = 3\]
Minkowski distances \(( \text { when } \lambda \rightarrow \infty )\) are:
\[d _ { M } ( 1,2 ) = \max ( | 1 - 1 | , | 3 - 2 | , | 1 - 1 | , | 2 - 2 | , | 4 - 1 | ) = 3\]
\[d _ { M } ( 1,3 ) = 2 \text { and } d _ { M } ( 2,3 ) = 1\]
- Calculate the Minkowski distance \(( \lambda = 1 , \lambda = 2 , \text { and } \lambda \rightarrow \infty \text { cases) }\) between the first and second objects.
- Calculate the Mahalanobis distance between the first and second objects.
- Minkowski distance is:
\[\lambda = 1 . \mathrm { d } _ { \mathrm { M } } ( 1,2 ) = | 2 - 10 | + | 3 - 7 | = 12\]
\[\lambda = \text{2. } \mathrm { d } _ { \mathrm { M } } ( 1,2 ) = \mathrm { d } _ { \mathrm { E } } ( 1,2 ) = \left( ( 2 - 10 ) ^ { 2 } + ( 3 - 7 ) ^ { 2 } \right) ^ { 1 / 2 } = 8.944\]
\[\lambda \rightarrow \infty . \mathrm { d } _ { \mathrm { M } } ( 1,2 ) = \max ( | 2 - 10 | , | 3 - 7 | ) = 8\]
- \[\lambda = \text{1 .} \operatorname { d_M } ( 1,2 ) = | 2 - 10 | + | 3 - 7 | = 12 . \lambda = \text{2 .} \operatorname { d_M } ( 1,2 ) = \operatorname { dE } ( 1,2 ) = ( ( 2 - 10 ) 2 + ( 3 - 7 ) 2 ) 1 / 2 = 8.944 . \lambda \rightarrow \infty\] . \(\operatorname { d_M } ( 1,2 ) = \max ( | 2 - 10 | , | 3 - 7 | ) = 8\) . Since \(\Sigma = \left( \begin{array} { l l } { 19 } & { 11 } \\ { 11 } & { 7 } \end{array} \right)\) we have \(\Sigma ^ { - 1 } = \left( \begin{array} { c c } { 7 / 12 } & { - 11 / 12 } \\ { - 11 / 12 } & { 19 / 12 } \end{array} \right)\) Mahalanobis distance is: \(d _ { M H } ( 1,2 ) = 2\)
- R code for Mahalanobis distance
Common Properties of Similarity Measures
Similarities have some well-known properties:
- s ( p , q ) = 1 (or maximum similarity) only if p = q ,
- s ( p , q ) = s ( q , p ) for all p and q , where s ( p , q ) is the similarity between data objects, p and q .
Similarity Between Two Binary Variables
The above similarity or distance measures are appropriate for continuous variables. However, for binary variables a different approach is necessary.
| q=1 | q=0 | |
|---|---|---|
| p=1 | n | n |
| p=0 | n | n |
Simple Matching and Jaccard Coefficients
- Simple matching coefficient \(= \left( n _ { 1,1 } + n _ { 0,0 } \right) / \left( n _ { 1,1 } + n _ { 1,0 } + n _ { 0,1 } + n _ { 0,0 } \right)\) .
- Jaccard coefficient \(= n _ { 1,1 } / \left( n _ { 1,1 } + n _ { 1,0 } + n _ { 0,1 } \right)\) .
Calculate the answers to the question and then click the icon on the left to reveal the answer.
Given data:
- p = 1 0 0 0 0 0 0 0 0 0
- q = 0 0 0 0 0 0 1 0 0 1
The frequency table is:
| q=1 | q=0 | |
|---|---|---|
| p=1 | 0 | 1 |
| p=0 | 2 | 7 |
Calculate the Simple matching coefficient and the Jaccard coefficient.
- Simple matching coefficient = (0 + 7) / (0 + 1 + 2 + 7) = 0.7.
- Jaccard coefficient = 0 / (0 + 1 + 2) = 0.
2.4 Visualization
To understand thousands of rows of data in a limited time there is no alternative to visual representation. The objective of visualization is to reveal hidden information through simple charts and diagrams. Visual representation of data is the first step toward data exploration and formulation of an analytical relationship among the variables. In a whirl of complex and voluminous data, visualization in one, two, and three-dimension helps data analysts to sift through data in a logical manner and understand the data dynamics. It is instrumental in identifying patterns and relationships among groups of variables. Visualization techniques depend on the type of variables. Techniques available to represent nominal variables are generally not suitable for visualizing continuous variables and vice versa. Data often contains complex information. It is easy to internalize complex information through visual mode. Graphs, charts, and other visual representations provide quick and focused summarization.
Tools for Displaying Single Variables
Histograms are the most common graphical tool to represent continuous data. On the horizontal axis, the range of the sample is plotted. On the vertical axis are plotted the frequencies or relative frequencies of each class. The class width has an impact on the shape of the histogram. The histograms in the previous section were drawn from a random sample generated from theoretical distributions. Here we consider a real example to construct histograms.
The dataset used for this purpose is the Wage data that is included in the ISLR package in R. A full description of the data is given in the package. The following R code produces the figure below which illustrates the distribution of wages for all 3000 workers.
- Sample R code for Distribution of Wage

The data is mostly symmetrically distributed but there is a small bimodality in the data which is indicated by a small hump towards the right tail of the distribution.
The data set contains a number of categorical variables one of which is Race. A natural question is whether the wage distribution is the same across Race. There are several libraries in R which may be used to construct histograms across levels of categorical variables and many other sophisticated graphs and charts. One such library is ggplot2. Details of the functionalities of this library will be given in the R code below.
In the following figures, histograms are drawn for each Race separately.
- Sample R code for Histogram of Wage by Race

Because of the huge disparity among the counts of the different races, the above histograms may not be very informative. Code for an alternative visual display of the same information is shown below, followed by the plot.
- Sample R code for Histogram of Wage by Race (Alternative)

The second type of histogram also may not be the best way of presenting all the information. However further clarity is seen in a small concentration at the right tail.
Boxplot is used to describe the shape of data distribution and especially to identify outliers. Typically an observation is an outlier if it is either less than Q 1 - 1.5 IQR or greater than Q 3 + 1.5 IQR, where IQR is the inter-quartile range defined as Q 3 - Q 1 . This rule is conservative and often too many points are identified as outliers. Hence sometimes only those points outside of [Q 1 - 3 IQR, Q 3 + 3 IQR] are only identified as outliers.
- Sample R code for Boxplot of Distribution of Wage

The boxplot of the Wage distribution clearly identifies many outliers. It is a reflection of the histogram depicting the distribution of Wage. The story is clearer from the boxplots drawn on the wage distribution for individual races. Here is the R code:
Here is the boxplot that results: * Sample R code for Boxplot Wage by Race

Tools for Displaying Relationships Between Two Variables
Scatterplot.
The most standard way to visualize relationships between two variables is a scatterplot. It shows the direction and strength of association between two variables but does not quantify it. Scatterplots also help to identify unusual observations. In the previous section (Section 1(b).2) a set of scatterplots are drawn for different values of the correlation coefficient. The data there is generated from a theoretical distribution of multivariate normal distribution with various values of the correlation parameter. Below is the R code used to obtain a scatterplot for these data:
The following is the scatterplot of the variables Age and Wage for the Wage data. * Sample R Code for Relationship of Age and Wage

It is clear from the scatterplot that the Wage does not seem to depend on Age very strongly. However, a set of points towards the top are very different from the rest. A natural follow-up question is whether Race has any impact on the Age-Wage dependency or the lack of it. Here is the R code and then the new plot:
- Sample R Code for Relationship of Age and Wage

We have noted before that the disproportionately high number of Whites in the data masks the effects of the other races. There does not seem to be any association between Age and Wage, controlling for Race.
Contour plot
This is useful when a continuous attribute is measured on a spatial grid. They partition the plane into regions of similar values. The contour lines that form the boundaries of these regions connect points with equal values. In spatial statistics, contour plots have a lot of applications.
Contour plots join points of equal probability. Within the contour lines concentration of bivariate distribution is the same. One may think of the contour lines as slices of a bivariate density, sliced horizontally. Contour plots are concentric; if they are perfect circles then the random variables are independent. The more oval-shaped they are, the farther they are from independence. Note the conceptual similarity in the scatterplot series in Sec 1.(b).2. In the following plot, the two disjoint shapes in the interior-most part indicate that a small part of the data is very different from the rest.
Here is the R code for the contour plot that follows:
- Sample R Code for Contour Plot of Age and Wage

Tools for Displaying More Than Two Variables
Scatterplot matrix.
Displaying more than two variables on a single scatterplot is not possible. A scatterplot matrix is one possible visualization of three or more continuous variables taken two at a time.
The data set used to display the scatterplot matrix is the College data that is included in the ISLR package. A full description of the data is given in the package. Here is the R code for the scatterplot matrix that follows:
- Sample R Code for Scatterplot Matrix of College Attributes

Parallel Coordinates
An innovative way to present multiple dimensions in the same figure is by using parallel coordinate systems. Each dimension is presented by one coordinate and instead of plotting coordinates at the right angle to one another, each coordinate is placed side-by-side. The advantage of such an arrangement is that many different continuous and discrete variables can be handled within a parallel coordinate system, but if the number of observations is too large, the profiles do not separate out from one another and patterns may be missed.
The illustration below corresponds to the Auto data from the ISLR package. Only 35 cars are considered but all dimensions are taken into account. The cars considered are different varieties of Toyota and Ford, categorized into two groups: produced before 1975 and produced in 1975 or after. The older models are represented by dotted lines whereas the newer cars are represented by dashed lines. The Fords are represented by blue color and Toyotas are represented by pink color. Here is the R code for the profile plot of this data that follows:
- Sample R Code for Profile Plot of Toyota and Ford Cars

The differences among the four groups are very clear from the figure. Early Ford models had 8 cylinders, were heavy, and had high horsepower and displacement. Naturally, they had low MPG and less time to accelerate. No Toyota belonged to this category. All Toyota cars are built after 1975, have 4 cylinders (one exception only) and MPG performance belongs to the upper half of the distribution. Note that only 35 cars are compared in the profile plot. Hence each car can be followed over all the attributes. However had the number of observations been higher, the distinction among the profiles would have been lost and the plot would not be informative.
Interesting Multivariate Plots
Following are some interesting visualization of multivariate data. In Star Plot , stars are drawn according to rules as defined by their characteristics. Each axis represents one attribute and the solid lines represent each item’s value on that attribute. All attributes of the observations are possible to be represented; however, for the sake of clarity on the graph only 10 attributes are chosen.
Again, the starplot follows the R code for generating the plot:
- Sample R Code for Starplot of College Data

Another interesting plot technique with multivariate data is Chernoff Face where attributes of each observation are used to draw different features of the face. A comparison of 30 colleges and universities from the College dataset is compared below.
Again, R code and then the plot follows:
- Sample R Code for Comparison of Colleges and Universities

For comparison of a small number of observations on up to 15 attributes, Chernoff’s face is a useful technique. However, whether two items are more similar or less, depends on interpretation.
2.5 R Scripts
This course requires a fair amount of R coding. The textbook takes the reader through R codes relevant for the chapter in a step-by-step manner. Sample R codes are also provided in the Visualization section. In this section, a brief introduction is given on a few of the important and useful features of R.
Introductions to R are available at Statistical R Tutorials and Cran R Project . There are many other online resources available for R. R users’ groups are thriving and highly communicative. A few additional resources are mentioned in the Course Syllabus.
One of the most important features of R is its libraries. They are freely downloadable from CRAN site. It is not possible to make a list of ALL or even MOST R packages. The list is ever changing as R users community is continuously building and refining the available packages. The link below is a good starting point for a list of packages for data manipulation and visualization.
R Studio Useful Packages
R Library: ggplot2
R has many packages and plotting options for data visualization but possibly none of them are able to produce as beautiful and as customizable statistical graphics as ggplot2 does. It is unlike most other graphics packages because it has a deep underlying grammar based on the Grammar of Graphics (Wilkinson, 2005). It is composed of a set of independent components that can be composed in many different ways. This makes ggplot2 very powerful because the user is not limited to a set of pre-specified graphics. The plots can be built up iteratively and edited later. The package is designed to work in a layered fashion, starting with a layer showing the raw data and then adding layers of annotations and statistical summaries.
The grammar of graphics is an answer to a question: what is a statistical graphic?
In brief, the grammar tells us that a statistical graphic is a mapping from data to aesthetic attributes (color, shape, size) of geometric objects (points, lines, bars). The plot may also contain statistical transformations of the data and is drawn on a specific coordinate system. Faceting can be used to generate the same plot for different subsets of the dataset. It is the combination of these independent components that make up a graphic.
A brief description of the main components are as below:
- The data and a set of aesthetic mappings describe how variables in the data are mapped to various aesthetic attributes
- Geometric objects, geoms for short, represent what is actually on the plot: points, lines, polygons, etc.
- Statistical transformations, stats for short, summarise data in many useful ways. For example, binning and counting observations to create a histogram, or summarising a 2d relationship with a linear model. Stats are optional but very useful.
- A facet ing specification describes how to break up the data into subsets and how to display those subsets as small multiples. This is also known as conditioning or latticing/trellising.
The basic command for plotting is qplot(X, Y, data = <data name>) (quick plot!). Unlike the most common plot() command, qplot() can be used for producing many other types of graphics by varying geom() . Examples of a few common geom() are given below.
- geom = “point” is the default
- geom = “smooth” fits a smoother to the data and displays the smooth and its standard error
- geom = “boxplot” produces a box-and-whisker plot to summarise the distribution of a set of points
For continuous variables
- geom = “histogram” draws a histogram
- geom = “density” draws a density plot
For discrete variables
- geom = “bar” produces a bar chart.
Aesthetics and faceting are two important features of ggplot2. Color, shape, size and other aesthetic arguments are used if observations coming from different subgroups are plotted on the same graph. Faceting takes an alternative approach: It creates tables of graphics by splitting the data into subsets and displaying the same graph for each subset in an arrangement that facilitates comparison.
From Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis , Springer.
Markdown is an extremely useful facility in R which lets a user incorporate R codes and outputs directly in a document. For a comprehensive knowledge on Markdown and how to use it, you may consult R Markdown in the course STAT 485.
Source Code

Exploratory data analysis (EDA) is used by data scientists to analyze and investigate data sets and summarize their main characteristics, often employing data visualization methods.
EDA helps determine how best to manipulate data sources to get the answers you need, making it easier for data scientists to discover patterns, spot anomalies, test a hypothesis, or check assumptions.
EDA is primarily used to see what data can reveal beyond the formal modeling or hypothesis testing task and provides a provides a better understanding of data set variables and the relationships between them. It can also help determine if the statistical techniques you are considering for data analysis are appropriate. Originally developed by American mathematician John Tukey in the 1970s, EDA techniques continue to be a widely used method in the data discovery process today.
Learn how to leverage the right databases for applications, analytics and generative AI.
Register for the ebook on generative AI
The main purpose of EDA is to help look at data before making any assumptions. It can help identify obvious errors, as well as better understand patterns within the data, detect outliers or anomalous events, find interesting relations among the variables.
Data scientists can use exploratory analysis to ensure the results they produce are valid and applicable to any desired business outcomes and goals. EDA also helps stakeholders by confirming they are asking the right questions. EDA can help answer questions about standard deviations, categorical variables, and confidence intervals. Once EDA is complete and insights are drawn, its features can then be used for more sophisticated data analysis or modeling, including machine learning .
Specific statistical functions and techniques you can perform with EDA tools include:
- Clustering and dimension reduction techniques, which help create graphical displays of high-dimensional data containing many variables.
- Univariate visualization of each field in the raw dataset, with summary statistics.
- Bivariate visualizations and summary statistics that allow you to assess the relationship between each variable in the dataset and the target variable you’re looking at.
- Multivariate visualizations, for mapping and understanding interactions between different fields in the data.
- K-means Clustering is a clustering method in unsupervised learning where data points are assigned into K groups, i.e. the number of clusters, based on the distance from each group’s centroid. The data points closest to a particular centroid will be clustered under the same category. K-means Clustering is commonly used in market segmentation, pattern recognition, and image compression.
- Predictive models, such as linear regression, use statistics and data to predict outcomes.
There are four primary types of EDA:
- Univariate non-graphical. This is simplest form of data analysis, where the data being analyzed consists of just one variable. Since it’s a single variable, it doesn’t deal with causes or relationships. The main purpose of univariate analysis is to describe the data and find patterns that exist within it.
- Stem-and-leaf plots, which show all data values and the shape of the distribution.
- Histograms, a bar plot in which each bar represents the frequency (count) or proportion (count/total count) of cases for a range of values.
- Box plots, which graphically depict the five-number summary of minimum, first quartile, median, third quartile, and maximum.
- Multivariate nongraphical: Multivariate data arises from more than one variable. Multivariate non-graphical EDA techniques generally show the relationship between two or more variables of the data through cross-tabulation or statistics.
- Multivariate graphical: Multivariate data uses graphics to display relationships between two or more sets of data. The most used graphic is a grouped bar plot or bar chart with each group representing one level of one of the variables and each bar within a group representing the levels of the other variable.
Other common types of multivariate graphics include:
- Scatter plot, which is used to plot data points on a horizontal and a vertical axis to show how much one variable is affected by another.
- Multivariate chart, which is a graphical representation of the relationships between factors and a response.
- Run chart, which is a line graph of data plotted over time.
- Bubble chart, which is a data visualization that displays multiple circles (bubbles) in a two-dimensional plot.
- Heat map, which is a graphical representation of data where values are depicted by color.
Some of the most common data science tools used to create an EDA include:
- Python: An interpreted, object-oriented programming language with dynamic semantics. Its high-level, built-in data structures, combined with dynamic typing and dynamic binding, make it very attractive for rapid application development, as well as for use as a scripting or glue language to connect existing components together. Python and EDA can be used together to identify missing values in a data set, which is important so you can decide how to handle missing values for machine learning.
- R: An open-source programming language and free software environment for statistical computing and graphics supported by the R Foundation for Statistical Computing. The R language is widely used among statisticians in data science in developing statistical observations and data analysis.
For a deep dive into the differences between these approaches, check out " Python vs. R: What's the Difference? "
Use IBM Watson® Studio to determine whether the statistical techniques that you are considering for data analysis are appropriate.
Learn the importance and the role of EDA and data visualization techniques to find data quality issues and for data preparation, relevant to building ML pipelines.
Learn common techniques to retrieve your data, clean it, apply feature engineering, and have it ready for preliminary analysis and hypothesis testing.
Train, validate, tune and deploy generative AI, foundation models and machine learning capabilities with IBM watsonx.ai, a next-generation enterprise studio for AI builders. Build AI applications in a fraction of the time with a fraction of the data.

IMAGES
VIDEO
COMMENTS
Exploratory data analysis (EDA) is an essential step in any research analysis. The. primary aim with exploratory analysis is to examine the data for distribution, outliers and anomalies to direct ...
This paper presents the results of an interview on exploratory data analysis with 18 analysts across academia and industry. We charac-terize common exploration goals: profiling (assessing data quality) and discovery (gaining new insights).
Exploratory data analysis (EDA) is an essential step in any research analysis. The primary aim with exploratory analysis is to examine the data for distribution, outliers and anomalies to direct specific testing of your hypothesis. It also provides tools for hypothesis generation by visualizing and understanding the data usually through ...
Exploratory Data Analysis (EDA) is a crucial step in the data analysis process that. aims to uncove r patterns, re lationships, and insights from raw data. It involves the use of various ...
Abstract Exploratory Data Analysis (EDA) is an important initial step for any knowledge discovery process, in which data scientists interactively explore unfamiliar datasets by issuing a sequence of analysis operations (e.g. filter, aggregation, and visualization). Since EDA is long known as a difficult task, requiring profound analytical skills, experience, and domain knowledge, a plethora of ...
To promote this balance in organizational science, rigorous inductive research aimed at phenomenon detection must be further encouraged. To this end, the present article discusses the logic and methods of exploratory data analysis (EDA), the mode of analysis concerned with discovery, exploration, and empirically detecting phenomena in data.
ABSTRACT Research and development in computer science and statistics have produced increasingly sophisticated software interfaces for interactive and exploratory analysis, optimized for easy pattern finding and data exposure. But design philosophies that emphasize exploration over other phases of analysis risk confusing a need for flexibility with a conclusion that exploratory visual analysis ...
How do analysis goals and context affect exploratory data analysis (EDA)? To investigate this question, we conducted semi-structured interviews with 18 data analysts. We characterize common exploration goals: profiling (assessing data quality) and discovery (gaining new insights). Though the EDA literature primarily emphasizes discovery, we observe that discovery only reliably occurs in the ...
Best practices in (teaching) data literacy, specifically Exploratory Data Analysis, remain an area of tacit knowledge until this day. However, with the increase in the amount of data and its importance in organisations, analysing data is becoming a much-needed skill...
Researchers must utilize exploratory data techniques to present findings to a target audience and create appropriate graphs and figures. Researchers can determine if outliers exist, data are missing, and statistical assumptions will be upheld by understanding data. Additionally, it is essential to comprehend these data when describing them in conclusions of a paper, in a meeting with ...
Later, we present a comprehensive survey of the recent advancements in the emerging field of exploratory data analysis. We investigate 50 academic and non-academic visual data exploration tools with respect to their utility in the six fundamental steps of the exploratory data analysis process.
Explore the latest full-text research PDFs, articles, conference papers, preprints and more on EXPLORATORY DATA ANALYSIS. Find methods information, sources, references or conduct a literature ...
This study uses exploratory data analysis to analyze financial accounting data, including balance sheets, income statements, and cash flow statement data.
Exploratory Data analysis (EDA) is one of the hidden and mundane tasks in analysis of Data, as a Model, Project or analysis is based on data, which is intuitive, extremely heterogenous and distorted in its form. (Data has become an integral part of every project, Model &) The analyzed data is more insightful for identifying and improving extremely critical business insights across the ...
Thorough knowledge of the structure of analyzed data allows to form detailed scientific hypotheses and research questions. The structure of data can be revealed with methods for exploratory data analysis. Due to multitude of available methods, selecting those which will work together well and facilitate data interpretation is not an easy task. In this work we present a well fitted set of tools ...
We believe that theories of a fuller integration of exploratory analysis with theories of inference can serve two purposes: it can facilitate the greater use of EDA in data science, and it can point to ways of making research and development in interactive statistical graphics more useful.
Correlation analysis is both popular and useful in a number of social networking research, particularly in the exploratory data analysis. In this paper, three well-known and often-used correlation co...
Exploratory Data Analysis (EDA) is a field of data analysis used to visually represent the knowledge embedded deep in the given data set. The technique is widely used to generate inferences from a given data set. Data set of current pandemic, the COVID-19 is widely made available by the standard dataset repository. EDA can be applied to these standard dataset to generate inferences. In this ...
The huge volume of data is used to make decision which is more accurate than intuition. Exploratory Data Analysis (EDA) detects mistakes, finds appropriate data, checks assumptions and determines the correlation among the explanatory variables. In the context, EDA is considered as analysing data that excludes inferences and statistical modelling.
Exploratory data analysis (EDA) is a data analysis approach that involves summarizing the main characteristics of the data and visualizing the data summary using appropriate representations [102].
Chapter 4 Exploratory Data Analysis. Chapter 4Exploratory D. ta AnalysisA rst look at the data.As mentioned in Chapter 1, exploratory data analysis or \EDA" is a critical rst step in an. lyzing the data from an experiment. H. detection of mistakes.
Exploratory Data Analysis (EDA) may also be described as data-driven hypothesis generation. Given a complex set of observations, often EDA provides the initial pointers towards various learning techniques. The data is examined for structures that may indicate deeper relationships among cases or variables.
Exploratory data analysis (EDA) is used by data scientists to analyze and investigate data sets and summarize their main characteristics, often employing data visualization methods. EDA helps determine how best to manipulate data sources to get the answers you need, making it easier for data scientists to discover patterns, spot anomalies, test ...
We have used exploratory data analysis (EDA) where data interpretations can be done in row and column format. We have used python for data analysis.
The analysis finds: • 83 per cent of all new migrants settled in a capital city metropolitan area. ... Research Papers. IPA Facts. Submissions. Parliamentary Research Briefs. Media Releases. Opinion. IPA Digital. Australia Censored. Australia's Future with Tony Abbott. The Great Books of Literature.